Difference between revisions of "Part:BBa K801100"
NoreenLouis (Talk | contribs) |
DaniekHoorn (Talk | contribs) |
||
| Line 59: | Line 59: | ||
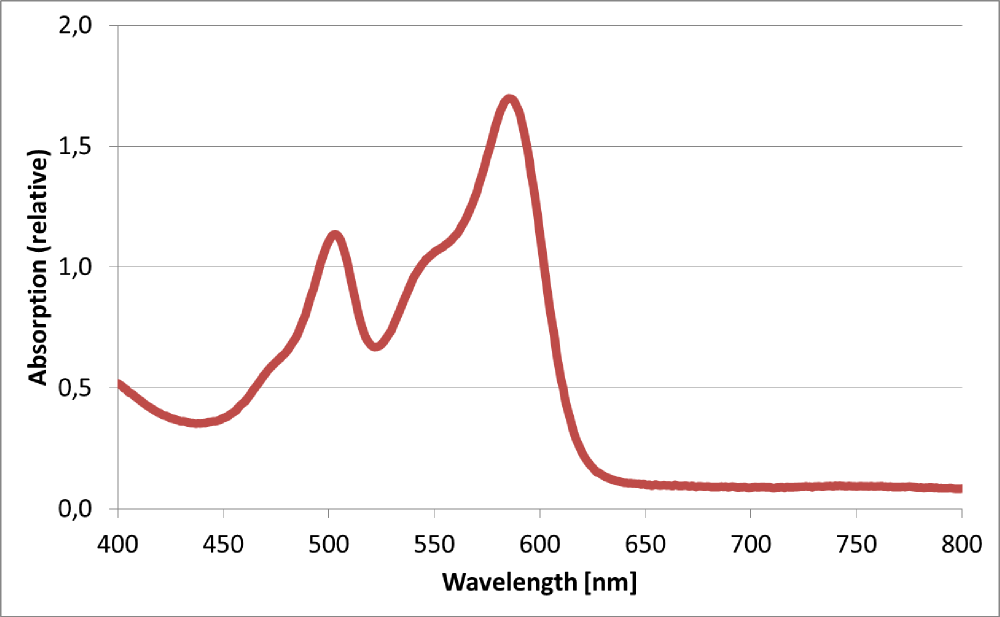
[[Image:TUM12_K801100spectrum.png |thumb|left|400x300px|Absorption spectrum of cell lysate expressing <partinfo>BBa_K801100</partinfo>]] | [[Image:TUM12_K801100spectrum.png |thumb|left|400x300px|Absorption spectrum of cell lysate expressing <partinfo>BBa_K801100</partinfo>]] | ||
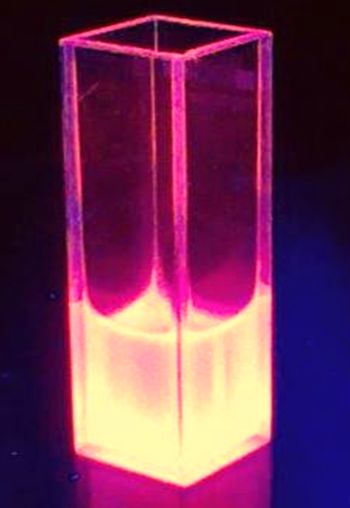
[[Image:TUM12_K801100fluorescence picture.jpg |thumb|left|160x300px|Cell lysate of <partinfo>BBa_K801100</partinfo>-expressing ''E. coli'' under UV illumination]] | [[Image:TUM12_K801100fluorescence picture.jpg |thumb|left|160x300px|Cell lysate of <partinfo>BBa_K801100</partinfo>-expressing ''E. coli'' under UV illumination]] | ||
| + | |||
| + | ==Improve== | ||
| + | The [https://2021.igem.org/Team:TU-Eindhoven/Improve iGEM team Eindhoven 2021] improved this part by removing the RBS (BBa_B0034) and by replacing this RBS by a sequence containing a 5'UTR and g10-L RBS ([https://parts.igem.org/Part:BBa_K3972006 BBa_K3972006]) to enhance expression of mRFP1. The addition of a 5’ untranslated region (5’-UTR) is already proven to increase expression of the consecutive gene, compared to the standard RBS BBa_B0034 [1]. The research paper of Volkenborn et. al confirms the importance of a 5’UTR sequence for translation initiation [2]. Part BBa_K3972006 is designed to optimally enhance protein expression in prokaryotes and it consists of translation-enhancing DNA, a 10-A-spacer, a g10-L RBS, and an AT-rich region. | ||
| + | Characterization methods and results can be found on part page [https://parts.igem.org/Part:BBa_K3972005 BBa_K3972005]. As can be seen on this page, brighter colonies were grown by <em>E. coli</em> Top 10 cells containing BBa_K3972005, compared to colonies grown in the same type of cells containing BBa_K801100. Therefore, looking at the grown colonies on the agar plates and the grown small cultures, it can carefully be claimed that faster mRFP1 expression occurs when the RBS (BBa_B0034) is replaced by a 5’UTR RBS (BBa_K3972006). However, further fluorescence intensity measurements need to be executed. | ||
| + | |||
| + | |||
| + | ==References== | ||
| + | [1] Takahashi S, Furusawa H, Ueda T, Okahata Y. Translation enhancer improves the ribosome liberation from translation initiation. J Am Chem Soc. 2013;135(35):13096–106. | ||
| + | |||
| + | [2] Volkenborn K, Kuschmierz L, Benz N, Lenz P, Knapp A, Jaeger K-E. The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis. Microb Cell Fact. 2020;19(1):154. | ||
Revision as of 08:54, 21 October 2021
RFP Coding Device - RFC10 and RFC25 compatible
This is an improved version of the RFP generating device BBa_J04450 designed by [http://openwetware.org/wiki/Davidson:Davidson_2005 Team Davidson 2005] that is now RFC10 and RFC25 compatible.
In 2012 the Registry of Standard Biological Parts defined BBa_J04450 as the standard shipping part that is required for submission of backbones, creating the need for a RFC25 compatible standard shipping part.
[http://2017.igem.org/Team:Grenoble-Alpes Team Grenoble-Alpes 2017] contributed to the characterisation of this part by testing the time of apparition of fluorescence as a signal of detection.
(--NoreenLouis 20:47, 26 October 2017 (UTC) )
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Composite part of the following BioBricks:
- BBa_R0010: Promoter (lacI regulated)
- BBa_B0034: RBS (Elowitz 1999)
- BBa_E1010: Red Fluorescent Protein from Discosoma striata
- BBa_B0015: Double Terminator
Functional parameters
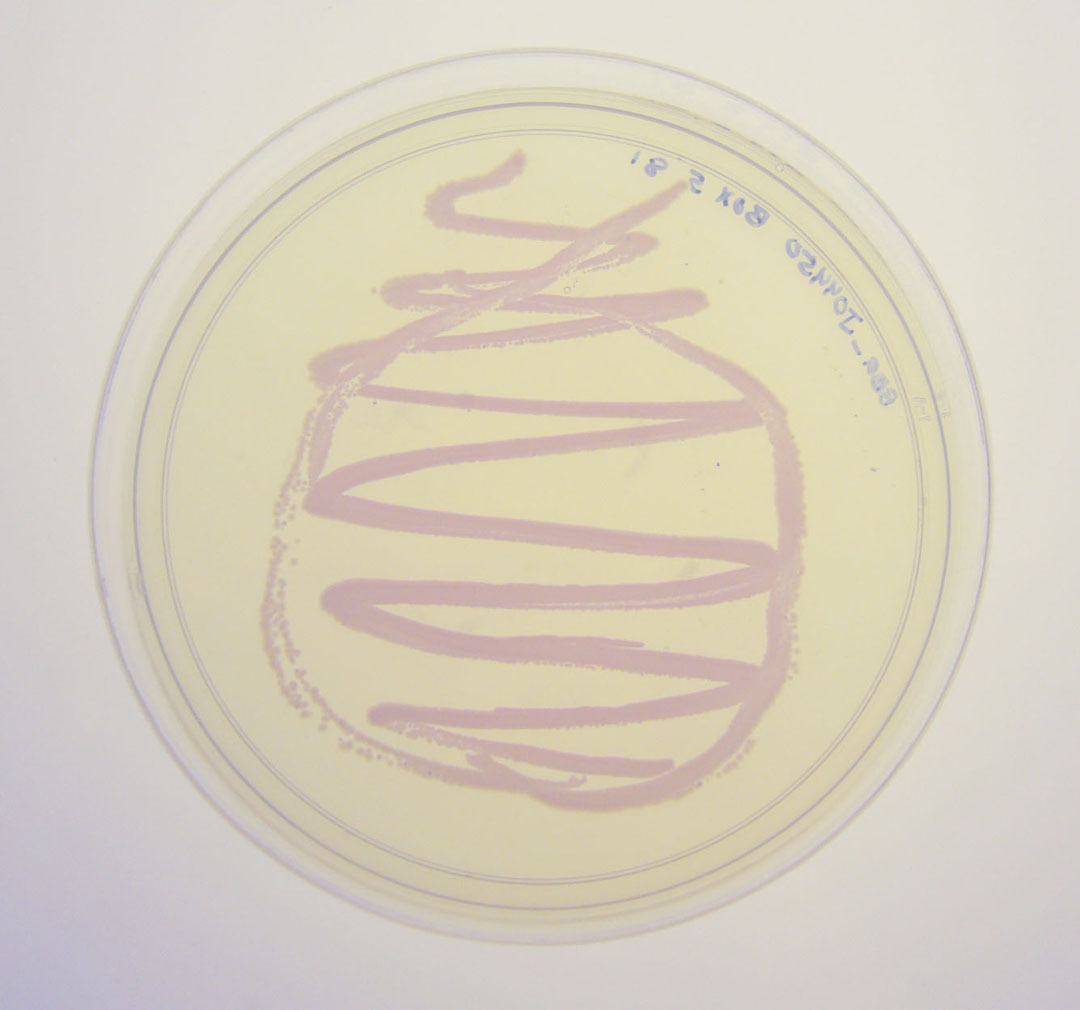
The colonies are clearly red in color under natural light after about 18 hours. Smaller colonies are visibly red under UV. The RFP part does not contain a degradation tag and the RBS is strong.
- LacI sensitive
- CAP sensitive
This part is commonly used, but can fail if the system contains LacI or CAP protein.
(--Meagan 15:39, 23 July 2009 (UTC))
Petri dish with BBa_J04450 visualized under non-UV lightbox
Petri dish with BBa_J04450 visualized under 254 nm wavelength UV lightbox
Colonies with and without BBa_J04450 on petri dish.
Usage as a cloning tool
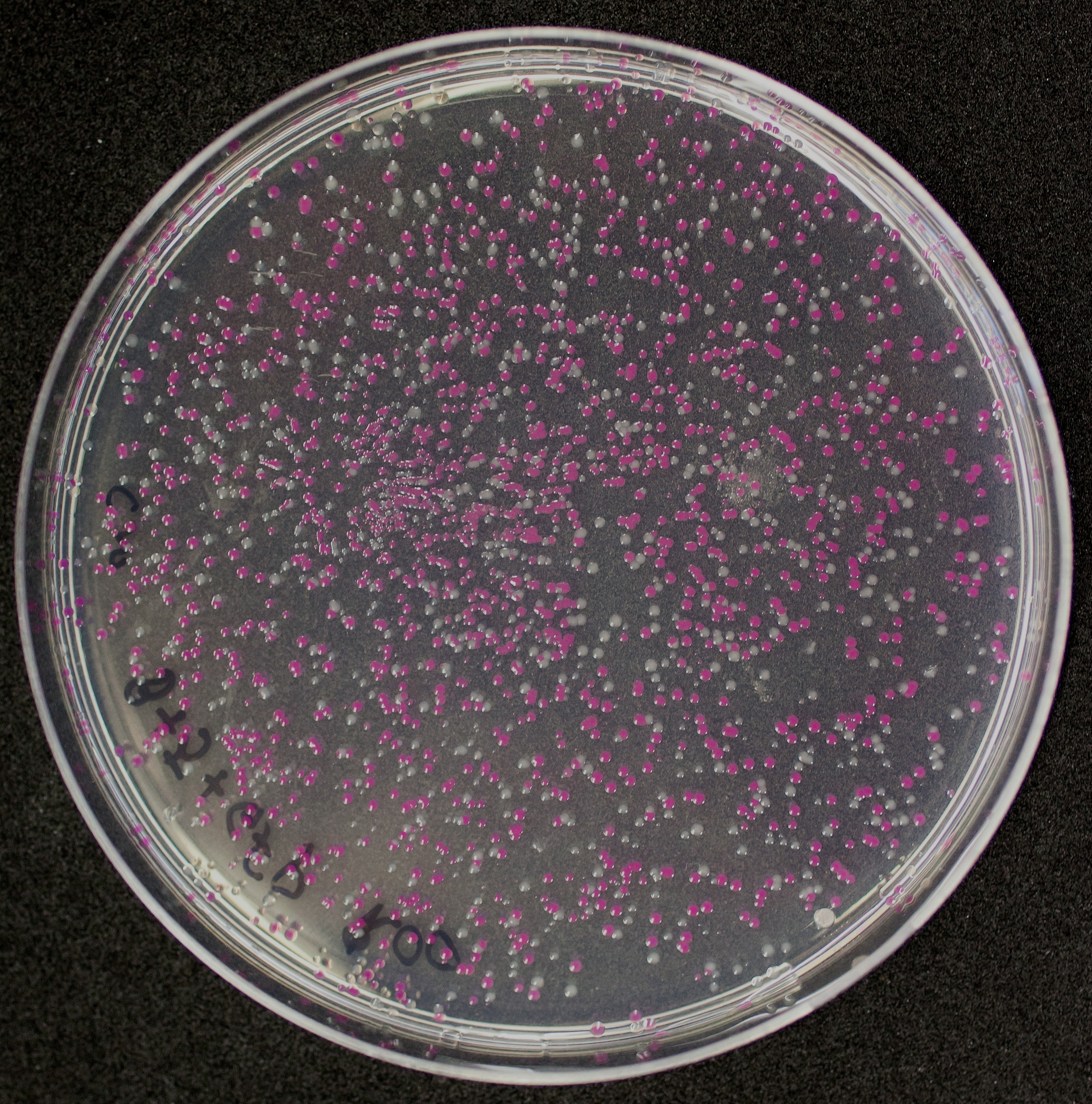
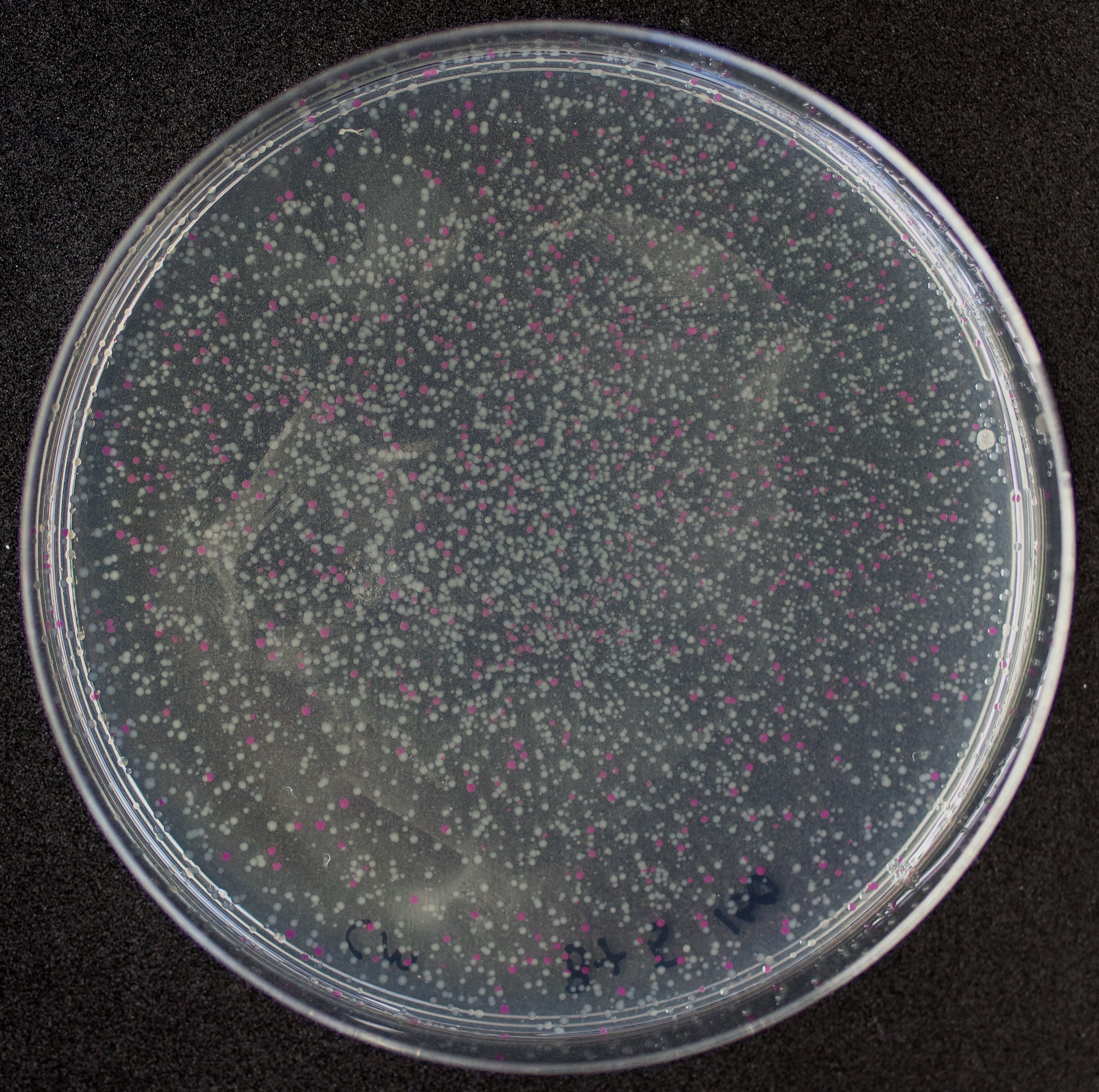
[http://2010.igem.org/Team:Groningen Team Groningen 2010] reports the usage of this part as a cloning tool. When ligating any part, or part assembly, into any standard backbone that contains this part, the non-restricted and single-restricted backbones that self-circularize will produce red colonies on rich media plates (we use TY). These undesired transformants can than be avoided in the screening for the correct construct. With this method, the backbone desired for a new construct does not need to be purified from agarose gel to decrease the amount of undesired tranformants caused by ligation of the original part present in the backbone. The amount of incorrect transformants depends, of course, on the ratio of backbone (mixed with BBa_J04450) vs. BioBrick insert, the size of the BioBrick insert, and whether the insert is an assembly of two BioBricks. The images below show two ligations with different efficiencies.
Extension of the standard compability to RFC10 and RFC25
[http://2012.igem.org/Team:TU_Munich Team TU_Munich 2012] extended the standard compability of the RFP coding device (BBa_J04450) to RFC10 and [http://hdl.handle.net/1721.1/45140 RFC25] by adding the NgoMIV and AgeI restriction sites into the prefix and suffix of this part. Additionally two AgeI restriction sites that were present in the generator itself were deleted.
This part may be used as a standard insert for RFC10 as well as [http://hdl.handle.net/1721.1/45140 RFC25] backbones. This improvement became necessary because insertion of BBa_J04450 into a RFC25 compatible backbone leads to the deletion of the desired [http://hdl.handle.net/1721.1/45140 RFC25] restriction sites that are needed for protein fusions.
Absorption spectrum from RFP1 produced by BBa_K801100
The BioBrick BBa_K801100 was characterized by expression of RFP1 (BBa_E1010) in E. coli XL1 blue. Cells were cultivated on a LB petri dish to guarantee sufficient oxygen supply. Afterward the cells were harvested and desintegrated using ultrasonic sound and cell debris was removed using centrifugation. Cell lysate in a cuvette is shown in the figure on the right. The absorption of the cell extract was measured in a photometer. The spectrum recorded is shown in the figure on the left in witch the RFP absorption peakt arroung 584 nm is present as it is described in literature.
Improve
The iGEM team Eindhoven 2021 improved this part by removing the RBS (BBa_B0034) and by replacing this RBS by a sequence containing a 5'UTR and g10-L RBS (BBa_K3972006) to enhance expression of mRFP1. The addition of a 5’ untranslated region (5’-UTR) is already proven to increase expression of the consecutive gene, compared to the standard RBS BBa_B0034 [1]. The research paper of Volkenborn et. al confirms the importance of a 5’UTR sequence for translation initiation [2]. Part BBa_K3972006 is designed to optimally enhance protein expression in prokaryotes and it consists of translation-enhancing DNA, a 10-A-spacer, a g10-L RBS, and an AT-rich region. Characterization methods and results can be found on part page BBa_K3972005. As can be seen on this page, brighter colonies were grown by E. coli Top 10 cells containing BBa_K3972005, compared to colonies grown in the same type of cells containing BBa_K801100. Therefore, looking at the grown colonies on the agar plates and the grown small cultures, it can carefully be claimed that faster mRFP1 expression occurs when the RBS (BBa_B0034) is replaced by a 5’UTR RBS (BBa_K3972006). However, further fluorescence intensity measurements need to be executed.
References
[1] Takahashi S, Furusawa H, Ueda T, Okahata Y. Translation enhancer improves the ribosome liberation from translation initiation. J Am Chem Soc. 2013;135(35):13096–106.
[2] Volkenborn K, Kuschmierz L, Benz N, Lenz P, Knapp A, Jaeger K-E. The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis. Microb Cell Fact. 2020;19(1):154.
(by [http://2012.igem.org/Team:TU_Munich Team TU_Munich 2012])