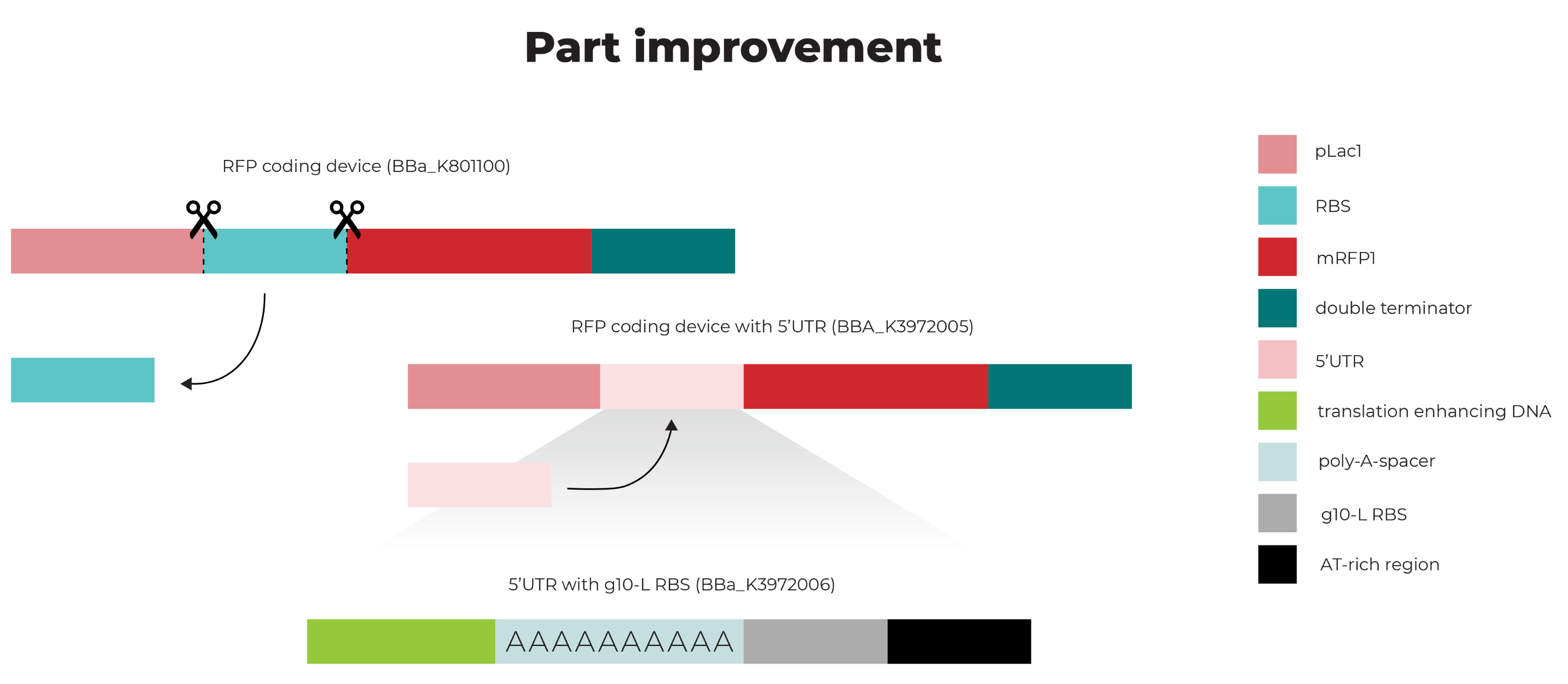
Part:BBa_K3972005
RFP coding device containing translation enhancing 5UTR
This part is an improved part of the RFP Coding Device (RFC10 and RFC25 compatible) BBa_K801100, which is improved by the replacement of the RBS BBa_B0034 by a translation enhancing 5’UTR containing a g10-L RBS (BBa_K3972006). This last part is based on a different 5’UTR sequence (BBa_K1758100).
Usage and Biology
The addition of a 5’UTR to an RBS increases protein expression by enhancing translation of the consecutive gene. The characterized 5’UTR contains a 10-A-spacer, which is used for the recruitment of the ribosome by recognition of the 5’ capped mRNA [1]. Furthermore, the sequence contains translation-enhancing DNA, a g10-L RBS, and an AT-rich region, which together replace the RBS (BBa_B0034) (Figure 1).
Figure 1. 5’UTR RFP coding device.
Characterization
Part BBa_K3972005 was ordered from IDT and was successfully ligated in a pSB1C3 plasmid, provided by iGEM in the 384-well plates. To characterize the E.coli optimized RFP coding device containing a 5’UTR (pSB1C3-5’UTR-RFP), different experiments were executed.
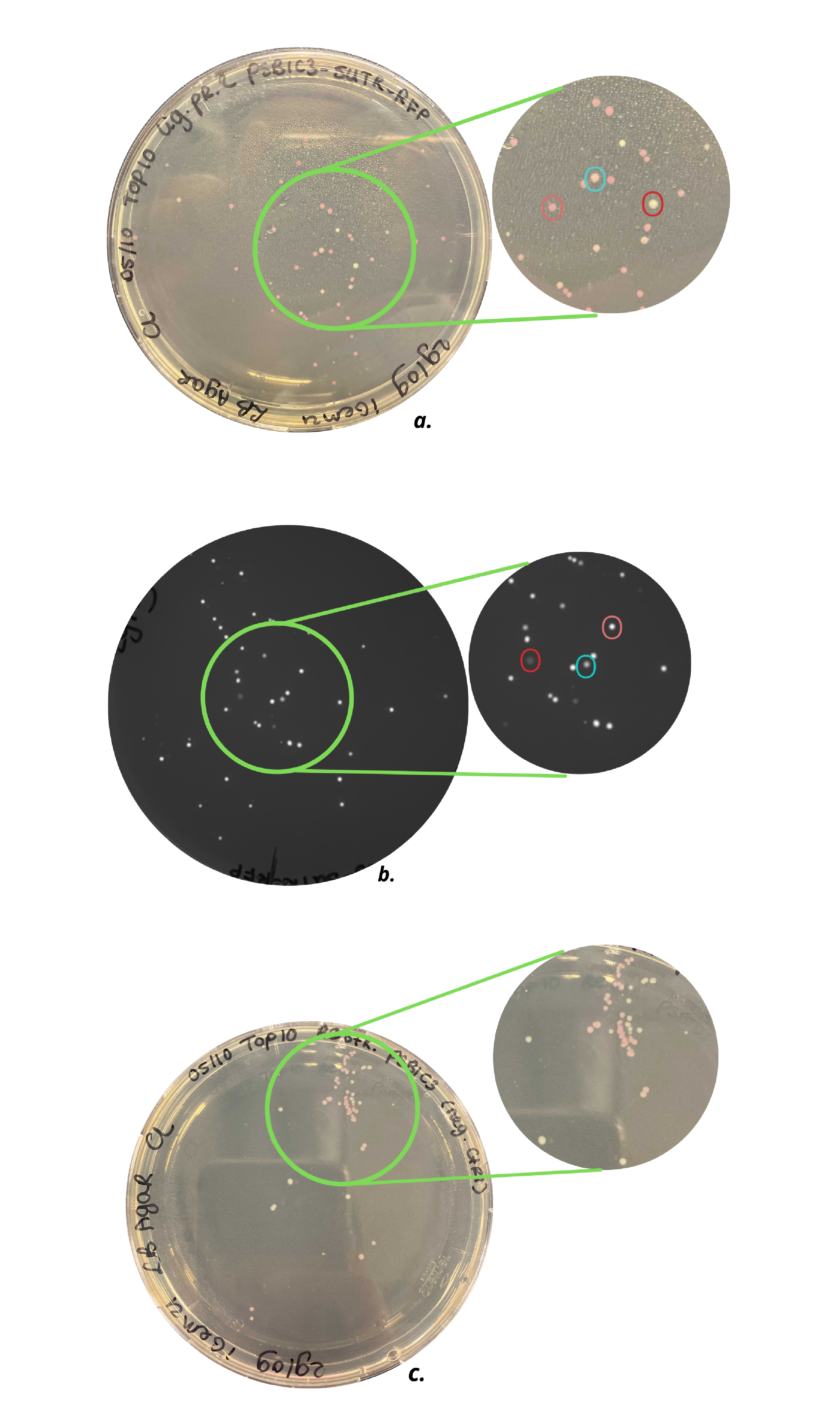
As can be seen in Figure 2, the pSB1C3-5’UTR-RFP plasmid was successfully transformed into E.coli Top10 cells. Additionally, the plasmids were successfully transformed in E.coli DH5alpha cells. Since the colonies of the Top10 cells showed more and brighter colonies, these colonies were used for further characterization. Figure 2c shows plates with colonies containing the pSB1C3 plasmid after restriction as a negative control. It can be concluded that not all pSB1C3 plasmids were properly restricted and that the colonies present on the agar plate most certainly contain unrestricted plasmids. The light pink colonies can be compared to the colonies present on the agar plate with colonies, containing the ligation product plasmid pSB1C3-5’UTR-RFP (Figure 2a). Three different colors of colonies can be observed on this plate, shown in the red, pink and light blue circle. The differences in fluorescence intensity of the colonies are shown in Figure 2b, which shows the same plate as in Figure 2b under UV light. The colony in the pink circle showed the highest fluorescence intensity and in addition, displayed the brightest pink color. From the brightest colonies, small cultures were made.
Figure 2. a) Top10 cells with pSB1C3-RFP plasmid, b) Top10 cells with pSB1C3-5’UTR-RFP plasmid under UV light, c) Top10 cells with a restricted pSB1C3-RFP plasmid (negative control). As can be seen in c) agar plate contains light pink and white colonies, which indicates not all pSB1C3-RFP plasmids are properly restricted. The agar plate of a) shows three types of colonies: white, light pink, and dark pink colonies, indicated with the red, blue, and pink circles, respectively. Agar plate b) shows the same colonies under UV light and an intensity difference can be observed.
The small culture of the original pSB1C3-RFP plasmid (BBa_K801100) and of the pSB1C3-5’UTR-RFP plasmid (BBa_K3972005) can be seen in Figure3. These small cultures were grown at the same time for approximately 22 hours. A proper color difference can be observed between the two cultures without induction, which already indicates faster mRFP1 expression by the bacteria containing the pSB1C3-5’UTR-RFP plasmid. The small cultures were used for large culture stocks, however, these large cultures did not grow properly. After 2.5 hours the OD600 of the cultures was only around 0.2, which can be explained by two different reasons. First of all, the small cultures were grown for 22 hours, so it is possible the bacterial cells reached the death phase[2]. Nonetheless, the large cultures were induced after 2.5 hours with three different concentrations of IPTG (0 mM, 0.5 mM and 2 mM) and grown overnight. The next day the large cultures showed no color difference and also fluorescence intensity measurements, using a Spark Plate Reader, didn’t show any applicable results. The induced large cultures were measured for 15 hours, every 15 minutes, and were excited at 584 nm. Due to time constraints the experiments could not be repeated.
Figure 3. Small cultures of wild type pSB1C3-RFP coding device plasmid (right) and improved pSB1C3-5'UTR-RFP (left) in E. coli Top 10 cells.
In conclusion, looking at the grown colonies on the agar plates and the grown small cultures, it can carefully be claimed that faster mRFP1 expression occurs when the RBS (BBa_B0034) is replaced by a 5’UTR RBS (BBa_K3972006). However, further fluorescence intensity measurements need to be executed.
References
[1] Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15(13):1593–612.
[2] Bailey R. Phases of the bacterial growth curve [Internet]. Thoughtco.com. [cited 2021 Oct 19]. Available from: https://www.thoughtco.com/bacterial-growth-curve-phases-4172692.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 809
Illegal AgeI site found at 921 - 1000COMPATIBLE WITH RFC[1000]
| emission | 607 nm |
| excitation | 584 nm |