Difference between revisions of "Protein domains/Overview"
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
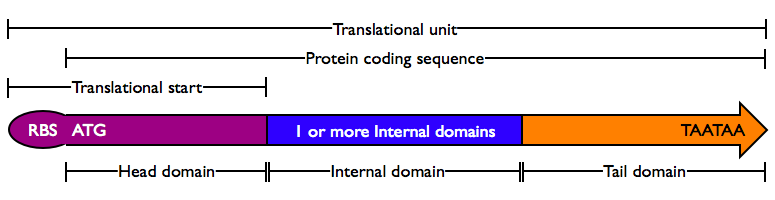
| − | Protein domains encode portions of proteins and can be assembled together to form translational units, a genetic part spanning translational initiation | + | Protein domains encode portions of proteins and can be assembled together to form translational units, a genetic part spanning from translational initiation (the RBS) to translational termination (the stop codon). |
[[Image:ProteinDomains.png|center]] | [[Image:ProteinDomains.png|center]] | ||
There are several different types of protein domains. | There are several different types of protein domains. | ||
| − | #'''Head Domain''': The Head domain consists of the | + | #'''Head Domain''': The Head domain consists of the start codon followed immediately by zero or more triplets specifiying an N-terminal tag, such as a protein export tag or lipoprotein binding tag. Head domains should begin with an <code>ATG</code> start codon and include codons 2 and 3 of the protein at a minimum. Examples of head domains include |
| − | #* | + | #*<code>ATG</code> start codon |
| − | #* | + | #*<code>ATG</code> start codon and codons 2-3 |
| − | #* | + | #*<code>ATG</code> start codon and signal sequence |
| − | #* | + | #*<code>ATG</code> start codon and affinity tag |
| − | #'''Internal Domains''': Protein domains consist of a series of codon triplets coding for an amino acid sequence without a start codon or stop codon. Multiple Domains can be fused | + | #'''Internal Domains''': Protein domains consist of a series of codon triplets coding for an amino acid sequence without a start codon or stop codon. Multiple Internal Domains can be fused. Examples of internal domains include |
| − | + | ||
#*DNA binding domains | #*DNA binding domains | ||
#*Dimerization domains | #*Dimerization domains | ||
#*Kinase domains | #*Kinase domains | ||
| + | #'''Special Internal Domains''': Short Domains with specific function may be separately categorized, but obey the same composition rules as normal internal domains. Examples of special internal domains include | ||
#*Linkers | #*Linkers | ||
#*Cleavage sites | #*Cleavage sites | ||
#*Inteins | #*Inteins | ||
| − | #'''Tail Domain''': The C-terminus of a coding region consists of zero or more triplet codons, followed by a pair of TAA stop codons. In the simplest case, the stop codons | + | #'''Tail Domain''': The C-terminus of a coding region consists of zero or more triplet codons, followed by a pair of TAA stop codons. In the simplest case, the stop codons terminate the protein with an Stop. More complex Tails may include degradation tags appropriate to the organism (i.e., with different degradation rates). Examples of Tail domain include |
| − | #* | + | #*<code>TAATAA</code> stop codons |
| − | #*A degradation tag followed by | + | #*A degradation tag followed by <code>TAATAA</code> stop codons |
| − | #*An affinity tag followed by | + | #*An affinity tag followed by <code>TAATAA</code> stop codon |
Unfortunately, the original BioBrick assembly standard, Assembly standard 10, does not support in-frame assembly of protein domains. (Assembly standard 10 creates an 8 bp scar between adjacent parts.) Therefore, it is recommended that you use an alternate approach to assemble protein domains together to make a translational unit. There are several possible approaches to [[Help:Protein domains/Assembly|assembling protein domains]] including direct synthesis (preferred because it creates no scars) as well as various assembly standards. Regardless of which standard you choose, we suggest that the resulting protein coding sequence or translational unit comply with the [[Help:Assembly standard 10|original BioBrick assembly standard]] so that your parts can be assembled with most of the parts in the Registry. | Unfortunately, the original BioBrick assembly standard, Assembly standard 10, does not support in-frame assembly of protein domains. (Assembly standard 10 creates an 8 bp scar between adjacent parts.) Therefore, it is recommended that you use an alternate approach to assemble protein domains together to make a translational unit. There are several possible approaches to [[Help:Protein domains/Assembly|assembling protein domains]] including direct synthesis (preferred because it creates no scars) as well as various assembly standards. Regardless of which standard you choose, we suggest that the resulting protein coding sequence or translational unit comply with the [[Help:Assembly standard 10|original BioBrick assembly standard]] so that your parts can be assembled with most of the parts in the Registry. | ||
| Line 30: | Line 30: | ||
| − | + | Note: Although most RBSs are currently specified as separate parts in the Registry, we are now moving to a new design in which the RBS and Head domain are combined into a single part termed a '''Translational start'''. The new design has the advantage of encapsulating both ribosome binding and translational initiation within a single part. Our working hypothesis is that the new design will reduce the likelihood of unexpected functional composition problems between the RBS and coding sequence. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | Although most RBSs are currently specified as separate parts in the Registry, we are now moving to a new design in which the RBS | + | |
Latest revision as of 22:57, 8 December 2009
Protein domains encode portions of proteins and can be assembled together to form translational units, a genetic part spanning from translational initiation (the RBS) to translational termination (the stop codon).
There are several different types of protein domains.
- Head Domain: The Head domain consists of the start codon followed immediately by zero or more triplets specifiying an N-terminal tag, such as a protein export tag or lipoprotein binding tag. Head domains should begin with an
ATGstart codon and include codons 2 and 3 of the protein at a minimum. Examples of head domains includeATGstart codonATGstart codon and codons 2-3ATGstart codon and signal sequenceATGstart codon and affinity tag
- Internal Domains: Protein domains consist of a series of codon triplets coding for an amino acid sequence without a start codon or stop codon. Multiple Internal Domains can be fused. Examples of internal domains include
- DNA binding domains
- Dimerization domains
- Kinase domains
- Special Internal Domains: Short Domains with specific function may be separately categorized, but obey the same composition rules as normal internal domains. Examples of special internal domains include
- Linkers
- Cleavage sites
- Inteins
- Tail Domain: The C-terminus of a coding region consists of zero or more triplet codons, followed by a pair of TAA stop codons. In the simplest case, the stop codons terminate the protein with an Stop. More complex Tails may include degradation tags appropriate to the organism (i.e., with different degradation rates). Examples of Tail domain include
TAATAAstop codons- A degradation tag followed by
TAATAAstop codons - An affinity tag followed by
TAATAAstop codon
Unfortunately, the original BioBrick assembly standard, Assembly standard 10, does not support in-frame assembly of protein domains. (Assembly standard 10 creates an 8 bp scar between adjacent parts.) Therefore, it is recommended that you use an alternate approach to assemble protein domains together to make a translational unit. There are several possible approaches to assembling protein domains including direct synthesis (preferred because it creates no scars) as well as various assembly standards. Regardless of which standard you choose, we suggest that the resulting protein coding sequence or translational unit comply with the original BioBrick assembly standard so that your parts can be assembled with most of the parts in the Registry.
Protein coding sequences should be as follows
GAATTC GCGGCCGC T TCTAG [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
Note: Although most RBSs are currently specified as separate parts in the Registry, we are now moving to a new design in which the RBS and Head domain are combined into a single part termed a Translational start. The new design has the advantage of encapsulating both ribosome binding and translational initiation within a single part. Our working hypothesis is that the new design will reduce the likelihood of unexpected functional composition problems between the RBS and coding sequence.