Difference between revisions of "Device:BBa D0001"
(New page: <div style="font-family: Arial; padding: 00px; width: 680px; border: 0px solid #000000;"> {{Template:Device page header}} {{Template:F2620 Characterization Navbar}} <div style="padding: 00...) |
|||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | <div style="font-family: Arial; padding: 00px; width: 680px; border: 0px solid #000000;"> | + | <div style="font-family: Arial; padding: 00px; width: 680px; border: 0px solid #000000; margin-left: auto; margin-right: auto"> |
{{Template:Device page header}} | {{Template:Device page header}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | {| | ||
| + | |'''Description''' | ||
| + | |'''Subparts''' | ||
| + | |- | ||
| + | |BBa_D0001 is a receiver device that can be used to sense the level of a cell-cell signaling molecule [[3OC6HSL|3OC<sub>6</sub>HSL]]. A transcription factor, LuxR (''<partinfo>C0062</partinfo>'') that is active in the presence of [[3OC6HSL|3OC<sub>6</sub>HSL]] is controlled by a TetR (''<partinfo>BBa_C0040</partinfo>'') regulated operator (''<partinfo>BBa_R0040</partinfo>''). ''Device input'' is [[3OC6HSL|3OC<sub>6</sub>HSL]]. ''Device output'' is [[PoPS]] from a LuxR-regulated operator. This device is known to work in many ''E. coli'' strains at a range of temperatures and in a variety of media types. | ||
| + | |||
| + | |<html><TABLE class='order_row' style='border: none;'border='0' cellpadding='0' cellspacing='0' color='#000000' bgcolor='#FFFFFF'><TR><TD style='font-size:90%; line-height:100%;text-align:center;'>tetR<br>R0040</TD><TD style='font-size:90%; line-height:100%;text-align:center;'>LuxR<br>I0462</TD><TD style='font-size:90%; line-height:100%;text-align:center;'>lux pR<br>R0062</TD><TR class = 'bar_row'><TD style='border: none;'><A href='/r/parts/partsdb/view.cgi?part_id=187'><img title='BBa_R0040 | ||
| + | promoter (tetR, negative)' src='https://parts.igem.org/images/partbypart/icon_regulatory.png' height='25' border=0></A><TD style='border: none;'><A href='/r/parts/partsdb/view.cgi?part_id=2681'><img title='BBa_I0462 | ||
| + | luxR Protein Generator' src='https://parts.igem.org/images/partbypart/icon_generator.png' height='25' border=0></A><TD style='border: none;'><A href='/r/parts/partsdb/view.cgi?part_id=192'><img title='BBa_R0062 | ||
| + | Promoter (luxR & HSL regulated -- lux pR)' src='https://parts.igem.org/images/partbypart/icon_regulatory.png' height='25' border=0></A></TABLE></html> | ||
| + | |} | ||
'''Performance'''<br> | '''Performance'''<br> | ||
| + | [[Image:BBa F2620TransferFunctionOct07.png|thumb|300px|right|'''Figure 1 - Transfer function of [[BBa_D0001]]. The data points represent the mean of 6 or 9 individual measurements. The corresponding error bars represent the 95% confidence interval in the mean of the independent measurements. The blue shaded region represents the range bounded by the lowest and highest outputs among the independent measurements at a given input level. The solid black curve was calculated by fitting a simple Hill function to the experimental measurements.''']] | ||
| + | [[Image:ActiviatedPromoterEquation.png|right|300px]] | ||
<center> | <center> | ||
{|{{Table}} | {|{{Table}} | ||
| Line 40: | Line 49: | ||
|[[Performance Stability|''Performance Stability'']]<br>(Low/High Input) | |[[Performance Stability|''Performance Stability'']]<br>(Low/High Input) | ||
|>92/>56 generations | |>92/>56 generations | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
</center> | </center> | ||
| − | |||
<div style="padding: 00px; width: 680px"> | <div style="padding: 00px; width: 680px"> | ||
'''Compatibility'''<br> | '''Compatibility'''<br> | ||
| Line 63: | Line 59: | ||
[[Help:Signalling|''Cell-Cell Signaling Systems:'']] Crosstalk with devices using [[3OC6HSL|3OC<sub>6</sub>HSL]], C6HSL, C7HSL, C8HSL, C10HSL. | [[Help:Signalling|''Cell-Cell Signaling Systems:'']] Crosstalk with devices using [[3OC6HSL|3OC<sub>6</sub>HSL]], C6HSL, C7HSL, C8HSL, C10HSL. | ||
</div> | </div> | ||
| − | + | ||
| + | ==Pages needed for a device== | ||
| + | *Main page | ||
| + | *Device Specification - experimental data and a model. Here is an example of a [http://openwetware.org/wiki/Registry_of_Standard_Biological_Models/Basic_Component_Models/Activated_Promoter model] for a biological device written in Cell ML. *User Comments and Usage Statistics | ||
| + | *Availability information for physical DNA | ||
Latest revision as of 16:14, 11 February 2009
Designed by: Francis Crick and Jim Watson
| AHL Receiver |
|
 |
| Description | Subparts | ||||||
| BBa_D0001 is a receiver device that can be used to sense the level of a cell-cell signaling molecule 3OC6HSL. A transcription factor, LuxR (BBa_C0062) that is active in the presence of 3OC6HSL is controlled by a TetR (BBa_C0040) regulated operator (BBa_R0040). Device input is 3OC6HSL. Device output is PoPS from a LuxR-regulated operator. This device is known to work in many E. coli strains at a range of temperatures and in a variety of media types. |
|
Performance
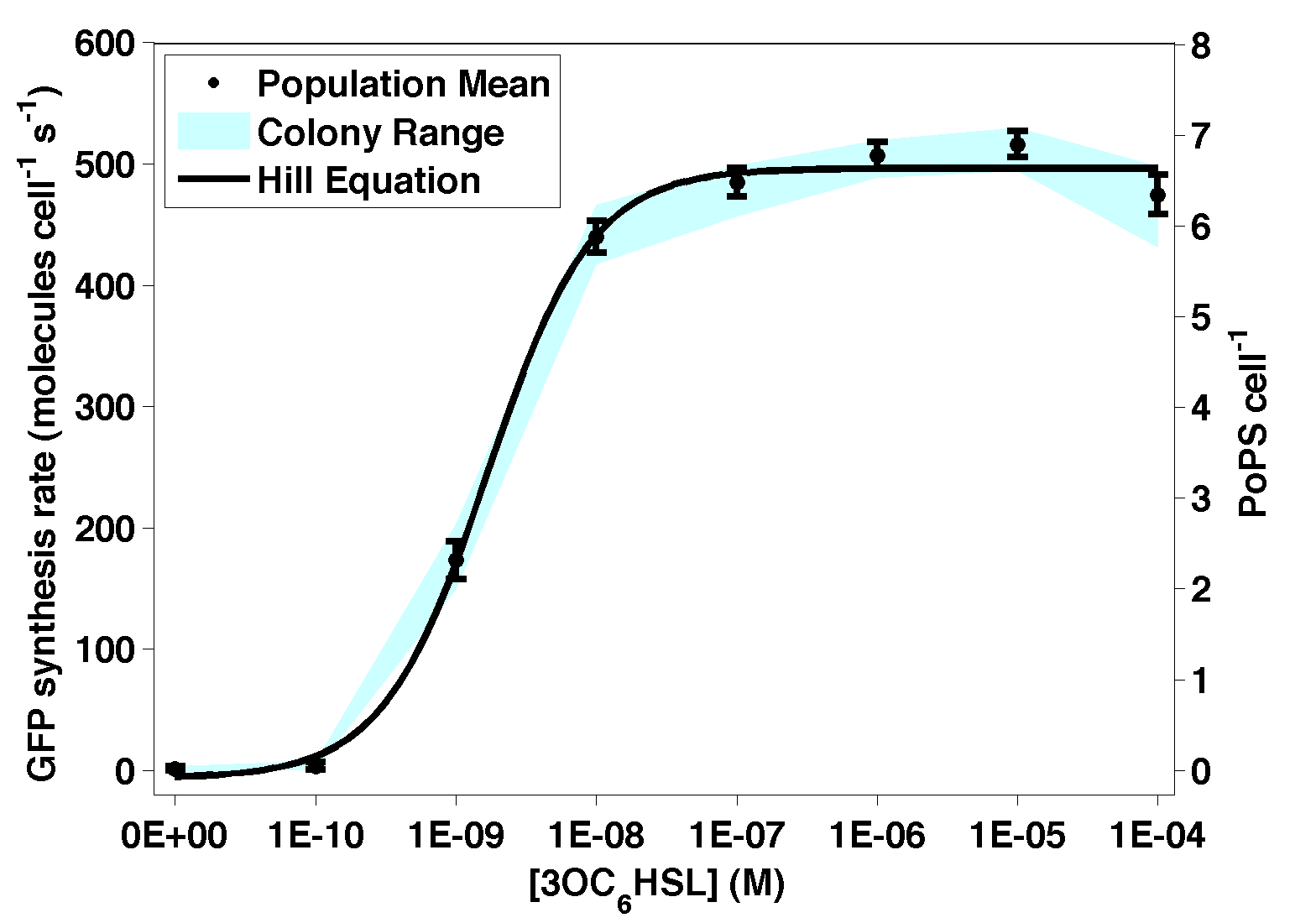
Figure 1 - Transfer function of BBa_D0001. The data points represent the mean of 6 or 9 individual measurements. The corresponding error bars represent the 95% confidence interval in the mean of the independent measurements. The blue shaded region represents the range bounded by the lowest and highest outputs among the independent measurements at a given input level. The solid black curve was calculated by fitting a simple Hill function to the experimental measurements.
| Experiment1 | Characteristic1 | Value1 |
|---|---|---|
| Transfer Function | Maximum Output | 6.6 PoPS cell-1 |
| Hill coefficient | 1.6 | |
| Switch Point | 1.5E-9 M 3OC6HSL, exogenous | |
| Response time: | <1 min | |
| Input compatibility | Strong response to | 3OC6HSL, C6HSL , C7HSL, 3OC8HSL, C8HSL |
| Weak response to | C4HSL, C10HSL, C12HSL | |
| Stability | Genetic Stability (Low/High Input) |
>92/>56 generations |
| Performance Stability (Low/High Input) |
>92/>56 generations |
Compatibility
Chassis: Device has been shown to work in BBa_V1000,BBa_V1001,BBa_V1002, E.Coli S30 Extract (data)
Plasmids: Device has been shown to work on pSB3K3 and pSB1A3
Devices: Device has been shown to work with BBa_E0430, BBa_E0434, BBa_E0240
Crosstalk with systems containing BBa_C0040.
Cell-Cell Signaling Systems: Crosstalk with devices using 3OC6HSL, C6HSL, C7HSL, C8HSL, C10HSL.
Pages needed for a device
- Main page
- Device Specification - experimental data and a model. Here is an example of a [http://openwetware.org/wiki/Registry_of_Standard_Biological_Models/Basic_Component_Models/Activated_Promoter model] for a biological device written in Cell ML. *User Comments and Usage Statistics
- Availability information for physical DNA
