Difference between revisions of "Part:BBa K3453111"
| (6 intermediate revisions by the same user not shown) | |||
| Line 66: | Line 66: | ||
The trigger sequences (Table 1) were placed under the control of the T7 promoter and followed by the strong SBa_000587 synthetic terminator for the T7 RNA polymerase and the resulting transcriptional units were synthesized and assembled in the high copy plasmid pSB1C3. | The trigger sequences (Table 1) were placed under the control of the T7 promoter and followed by the strong SBa_000587 synthetic terminator for the T7 RNA polymerase and the resulting transcriptional units were synthesized and assembled in the high copy plasmid pSB1C3. | ||
| − | For toehold switch characterisation we decided to use ''E. coli'' BL21 Star™(DE3) cells (Thermo Fisher Scientific), genotype F- ''ompT hsdS''B (rB-mB-) ''gal dcm rne131'' (DE3). As all BL21(DE3) ''E. coli'' strains, these cells contain the T7 RNA polymerase under control of the lacUV5 promoter and thus require IPTG to induce expression. The particularity of BL21 Star™(DE3) cells is that they contain a truncated version of the RNaseE gene (rne131) that leads to reduced level of mRNA degradation and thus increased RNA stability. | + | For toehold switch characterisation we decided to use ''E. coli'' BL21 Star™(DE3) cells (Thermo Fisher Scientific), genotype F- ''ompT hsdS''B (rB-mB-) ''gal dcm rne131'' (DE3). As all BL21(DE3) ''E. coli'' strains, these cells contain the T7 RNA polymerase under control of the lacUV5 promoter and thus require IPTG to induce expression. The particularity of BL21 Star™(DE3) cells is that they contain a truncated version of the RNaseE gene (''rne131'') that leads to reduced level of mRNA degradation and thus increased RNA stability. |
| + | |||
| + | [[Part:BBa_K3453111|BBa_K3453111]] was characterised both ''in vivo'' using ''E. coli'' as chassis and ''in vitro'' in a cell-free experimental set-up as a proof of concept for the final inexpensive, portable and easy-to-use rosewood biosensor. | ||
| + | |||
| + | ====''in vivo'' experimental characterization==== | ||
For fluorescence measurements, ''E. coli'' cells containing switch and trigger plasmids were first grown overnight in 96-deep-well plates containing 1 mL of LB medium supplemented with 5 µg/mL tetracycline and 17.5 µg/mL chloramphenicol, then diluted by 40x into similar media. Upon reaching early log-phase, cells were further diluted 20x in 100 µL of LB medium supplemented with 5 µg/mL tetracycline, 17.5 µg/mL chloramphenicol and 10 µM IPTG in an opaque wall 96-well polystyrene microplate, the COSTAR 96 (Corning). The plate was incubated at 37°C at 200 rpm and the sfGFP fluorescence (λexcitation 483 nm and λemission 530 nm) and OD600nm were measured every 10 min for 24 hours in a CLARIOstar (BMGLabtech) plate reader. | For fluorescence measurements, ''E. coli'' cells containing switch and trigger plasmids were first grown overnight in 96-deep-well plates containing 1 mL of LB medium supplemented with 5 µg/mL tetracycline and 17.5 µg/mL chloramphenicol, then diluted by 40x into similar media. Upon reaching early log-phase, cells were further diluted 20x in 100 µL of LB medium supplemented with 5 µg/mL tetracycline, 17.5 µg/mL chloramphenicol and 10 µM IPTG in an opaque wall 96-well polystyrene microplate, the COSTAR 96 (Corning). The plate was incubated at 37°C at 200 rpm and the sfGFP fluorescence (λexcitation 483 nm and λemission 530 nm) and OD600nm were measured every 10 min for 24 hours in a CLARIOstar (BMGLabtech) plate reader. | ||
Fluorescence values were normalised by OD600nm and, using the calibration curves presented on [https://2020.igem.org/Team:Evry_Paris-Saclay/Measurement the ‘Measurement’ page of our wiki], we converted the arbitrary units into Molecules of Equivalent FLuorescein (MEFL) / particle. | Fluorescence values were normalised by OD600nm and, using the calibration curves presented on [https://2020.igem.org/Team:Evry_Paris-Saclay/Measurement the ‘Measurement’ page of our wiki], we converted the arbitrary units into Molecules of Equivalent FLuorescein (MEFL) / particle. | ||
| − | The results presented in figures 2, 3 and 4 show that the rosewood DmMatK toehold switch 1.1 is functional and | + | The results presented in figures 2, 3 and 4 show that the rosewood DmMatK toehold switch 1.1 is functional and behaves as expected. When tested against its cognate trigger, this toehold switch showed fluorescence with a mean comparable to that of the positive controls (Figure 2) suggesting that we leveraged the full capacity of the expression cassette. The ON/OFF ratio of this toehold switch was greater than 100 (Figure 3), on the same order of performance as some of the best switches reported in literature. |
The results presented in Figure 2 show also that the sfGFP expression was readily detected in the positive controls and that the two RBS ([[Part:BBa_K2675017|BBa_K2675017]] and [[Part:BBa_K3453005|BBa_K3453005]]) have comparable strength. As expected, no fluorescence was detected in the negative controls as either the sfGFP gene was not present or the promoter and the RBS were absent. | The results presented in Figure 2 show also that the sfGFP expression was readily detected in the positive controls and that the two RBS ([[Part:BBa_K2675017|BBa_K2675017]] and [[Part:BBa_K3453005|BBa_K3453005]]) have comparable strength. As expected, no fluorescence was detected in the negative controls as either the sfGFP gene was not present or the promoter and the RBS were absent. | ||
| Line 88: | Line 92: | ||
Figure 4: Pictures of ''E. coli'' BL21 Star™(DE3) cells harbouring this part in the presence of the rosewood DmMatK triggers (Table 1). The negative control was performed with an empty pSB1C3 (no trigger). | Figure 4: Pictures of ''E. coli'' BL21 Star™(DE3) cells harbouring this part in the presence of the rosewood DmMatK triggers (Table 1). The negative control was performed with an empty pSB1C3 (no trigger). | ||
| + | |||
| + | ====''in vitro'' experimental characterization==== | ||
| + | An extract based cell-free system was prepared following Noireaux’s lab protocol [2] using the ''E. coli'' BL21 Star™(DE3) cells (Thermo Fisher Scientific). As our constructs are all placed under the control of T7 promoter, the T7 RNA polymerase expression was induced by adding 1 mM of IPTG to the 2YTP growth media during the production culture. Mg glutamate, K glutamate and DTT concentrations were optimized in order to have the highest efficiency of protein production achieved by the cell-free preparation. | ||
| + | Sensor plasmids containing the toehold switch sequence in front of the sfGFP gene in pSB3T5 backbone were purified in large quantities from 300 mL LB + 10 µg/mL tetracycline cultures followed by a MaxiPrep (Macherey Nagel) plasmid extraction. | ||
| + | |||
| + | The trigger RNA was produced from a PCR reamplified template using the TranscriptAid T7 (Thermo Fisher Scientific) transcription kit. Template trigger DNA was digested using the DNAse enzyme provided in the same kit and after a RNA purification step using the Monarch® RNA Cleanup Kit (New England Biolabs) the integrity and the size of the RNA transcript was confirmed by running an aliquot of the final product on a 2.5% agarose gel. All these steps were carried in an RNAse free environment with a specific attention paid to the avoidance of external RNAse contaminations. | ||
| + | |||
| + | Each of the sensors was tested in triplicate against water and their cognate triggers. The final DNA/RNA concentrations were 5 nM and 2 µM for switches and triggers respectively. Note that the switch concentrations correspond to DNA concentrations while the trigger concentrations correspond to RNA concentrations, thus we expect similar concentrations after transcription. | ||
| + | |||
| + | Reactions were run in an opaque wall 384-well polystyrene microplate (Nunc 242764) in a final volume of 15 µL constituted by 5.33 µL cell-free extract, 6.33 µL buffer and 3.33 µL of water, DNA and RNA. The plate was incubated at 30°C and the sfGFP fluorescence (λexcitation 485 nm and λemission 528 nm) wes measured every 5 min for 8 hours in a Synergy HTX (BioTek) plate reader. An endpoint measurement was extracted at 45 minutes to compare the fluorescence of the sensor with and without addition of the trigger RNA in a fast output setup. Fluorescence values were converted into Molecules of Equivalent FLuorescein (MEFL) using the calibration curves presented on the [https://2020.igem.org/Team:Evry_Paris-Saclay/Measurement the ‘Measurement’ page of our wiki]. | ||
| + | |||
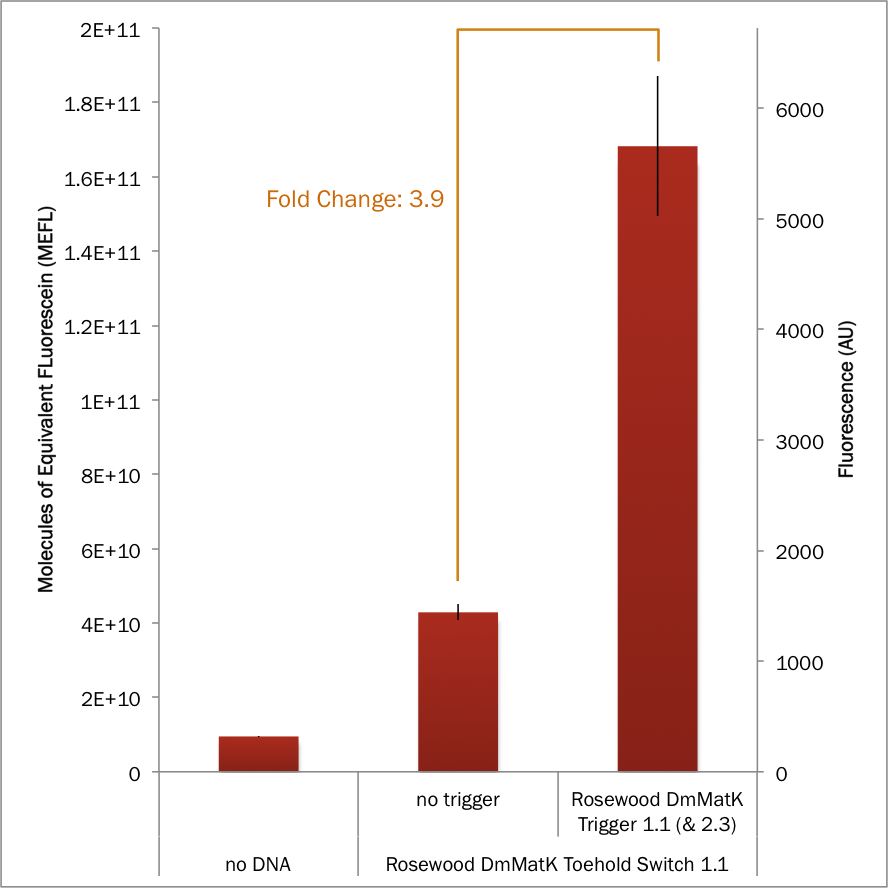
| + | When tested with its cognate trigger in the cell-free platform, [[Part:BBa_K3453111|BBa_K3453111]] showed a positive behavior. In the presence of the trigger the fluorescence signal was strongly increased compared to the signal obtained in the absence of the trigger (Figure 5), the fluorescence fold changes reaching a significant level. | ||
| + | |||
| + | |||
| + | [[File: T--Evry_Paris-Saclay--Cell-free_p35_DmMatK1-1.png|center|400px]] | ||
| + | |||
| + | Figure 5. Characterization of sfGFP expression controlled by the rosewood DmMatK toehold switch 1.1 and the rosewood DmMatK trigger 1.1 in an ''E. coli'' BL21 Star™(DE3) based cell-free system. The data and error bars are the mean and standard deviation of at least three measurements. Fold change is defined as the ratio of the average fluorescence of sensor + trigger cell extract vs the sensor + no trigger cell extract. | ||
===References=== | ===References=== | ||
[1] Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell (2014) 159, 925-939. | [1] Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell (2014) 159, 925-939. | ||
| + | |||
| + | [2] Sun ZZ, Hayes CA, Shin J, Caschera F, Murray RM, Noireaux V. Protocols for implementing an ''Escherichia coli'' based TX-TL cell-free expression system for synthetic biology. Journal of Visualized Experiments: JoVE (2013) e50762. | ||
Latest revision as of 00:38, 28 October 2020
Rosewood DmMatK Toehold Switch 1.1 with sfGFP-LVAtag
This part is an sfGFP-LVAtag (BBa_K2675006) expression cassette under the control of the Rosewood DmMatK Toehold Switch 1.1 (BBa K3453011) for sequence-based detection of the rosewood species Dalbergia maritima var. pubescens.
Usage and Biology
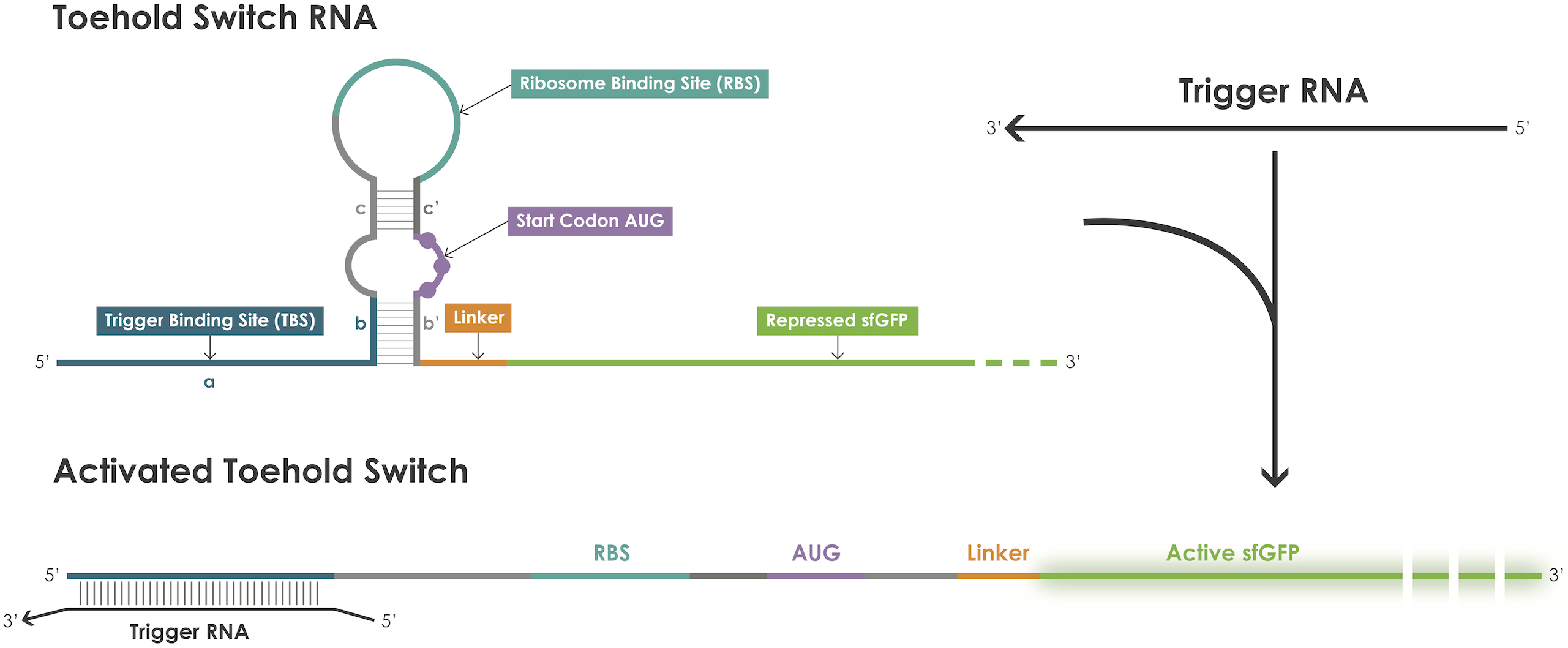
A toehold switch is an RNA–based device containing a ribosome binding site (RBS) and an ATG start codon embedded in the middle of a hairpin structure that blocks translation initiation [1]. The hairpin can be unfolded upon binding of a trigger RNA thereby exposing the RBS and the ATG start codon and thus permitting translation of the reporter protein (Figure 1).
Figure 1. Toehold switches principle.
In this composite part sfGFP-LVAtag (BBa_K2675006) was equipped by the synthetic Rosewood DmMatK Toehold Switch 1.1 (BBa K3453011) and placed under the control of the T7 promoter (BBa_K2150031) and of the strong SBa_000587 synthetic terminator (BBa_K3453000).
The corresponding trigger sequence of this toehold switch is the rosewood DmMatK Trigger 1.1 (Table 1).
| Table 1. Rosewood DmMatK triggers. | |||
|---|---|---|---|
| Label | Part number | Transcriptional units’ part numbers | Specificity |
| Rosewood DmMatK Trigger (Full) | BBa_K3453020 | BBa_K3453120 | all Rosewood DmMatK switches |
| Rosewood DmMatK Trigger 1.1 | BBa_K3453021 | BBa_K3453121 | Rosewood DmMatK switch 1.1, but also 2.3 |
| Rosewood DmMatK Trigger 1.3 | BBa_K3453022 | BBa_K3453122 | Rosewood DmMatK switch 1.3, but also 1.1 & 1.4 |
| Rosewood DmMatK Trigger 1.4 | BBa_K3453023 | BBa_K3453123 | Rosewood DmMatK switch 1.4, but also 1.1 & 2.3 |
| Rosewood DmMatK Trigger 2.1 | BBa_K3453024 | BBa_K3453124 | Rosewood DmMatK switch 2.1 |
| Rosewood DmMatK Trigger 2.2 | BBa_K3453025 | BBa_K3453125 | Rosewood DmMatK switch 2.2 |
| Rosewood DmMatK Trigger 2.3 | BBa_K3453026 | BBa_K3453126 | Rosewood DmMatK switch 2.3 |
BBa_K3453111 was assembled by Golden Gate in the low copy plasmid pSB3T5. For this, a fragments containing the BBa_K3453011 was synthesized flanked by BbsI type IIS restriction sites, and then inserted into BBa_K3453103, a platform in which a Golden Gate adapter with BbsI sites is present upstream of the sfGFP-LVAtag.
The trigger sequences (Table 1) were placed under the control of the T7 promoter and followed by the strong SBa_000587 synthetic terminator for the T7 RNA polymerase and the resulting transcriptional units were synthesized and assembled in the high copy plasmid pSB1C3.
For toehold switch characterisation we decided to use E. coli BL21 Star™(DE3) cells (Thermo Fisher Scientific), genotype F- ompT hsdSB (rB-mB-) gal dcm rne131 (DE3). As all BL21(DE3) E. coli strains, these cells contain the T7 RNA polymerase under control of the lacUV5 promoter and thus require IPTG to induce expression. The particularity of BL21 Star™(DE3) cells is that they contain a truncated version of the RNaseE gene (rne131) that leads to reduced level of mRNA degradation and thus increased RNA stability.
BBa_K3453111 was characterised both in vivo using E. coli as chassis and in vitro in a cell-free experimental set-up as a proof of concept for the final inexpensive, portable and easy-to-use rosewood biosensor.
in vivo experimental characterization
For fluorescence measurements, E. coli cells containing switch and trigger plasmids were first grown overnight in 96-deep-well plates containing 1 mL of LB medium supplemented with 5 µg/mL tetracycline and 17.5 µg/mL chloramphenicol, then diluted by 40x into similar media. Upon reaching early log-phase, cells were further diluted 20x in 100 µL of LB medium supplemented with 5 µg/mL tetracycline, 17.5 µg/mL chloramphenicol and 10 µM IPTG in an opaque wall 96-well polystyrene microplate, the COSTAR 96 (Corning). The plate was incubated at 37°C at 200 rpm and the sfGFP fluorescence (λexcitation 483 nm and λemission 530 nm) and OD600nm were measured every 10 min for 24 hours in a CLARIOstar (BMGLabtech) plate reader. Fluorescence values were normalised by OD600nm and, using the calibration curves presented on the ‘Measurement’ page of our wiki, we converted the arbitrary units into Molecules of Equivalent FLuorescein (MEFL) / particle.
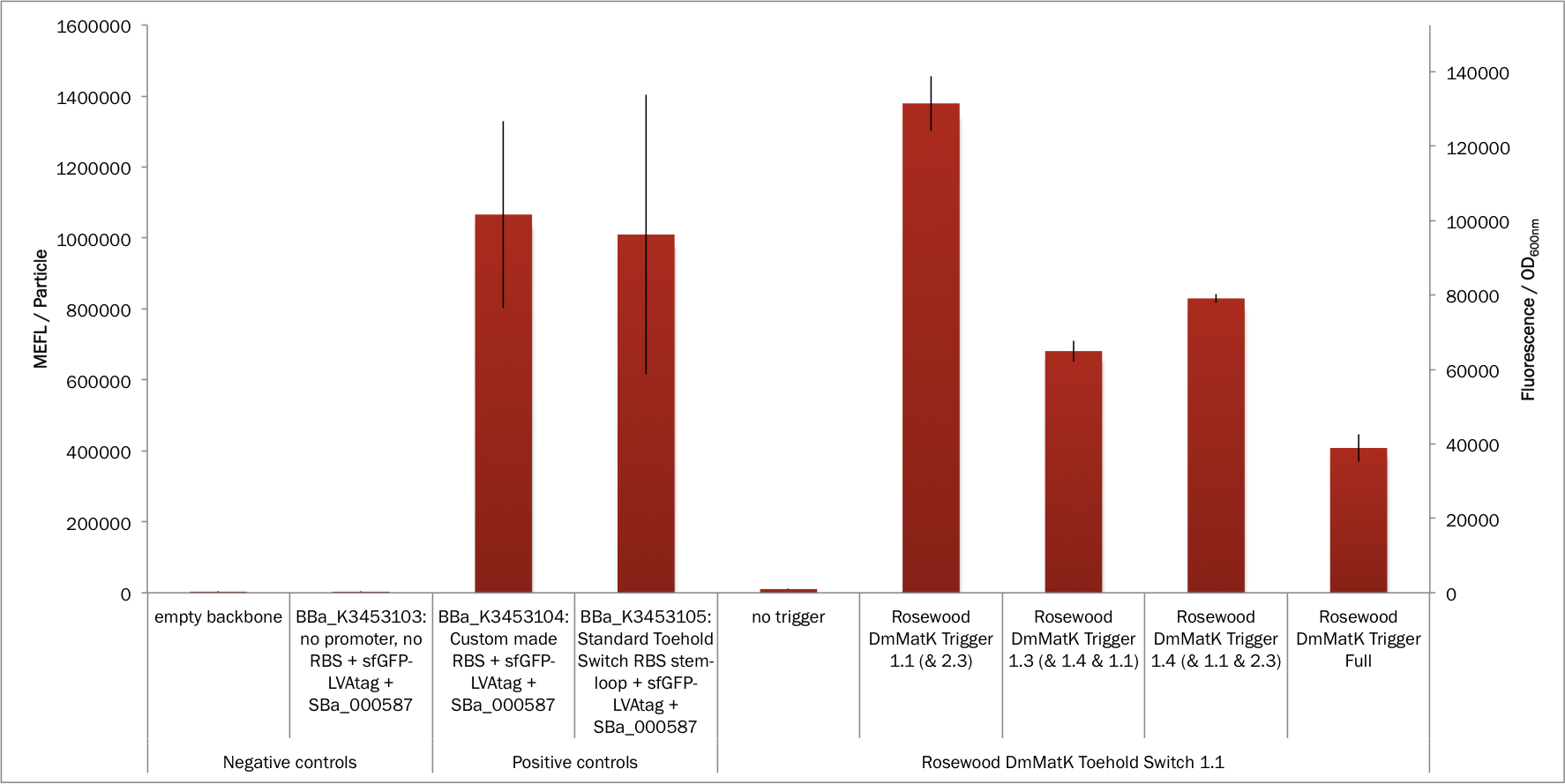
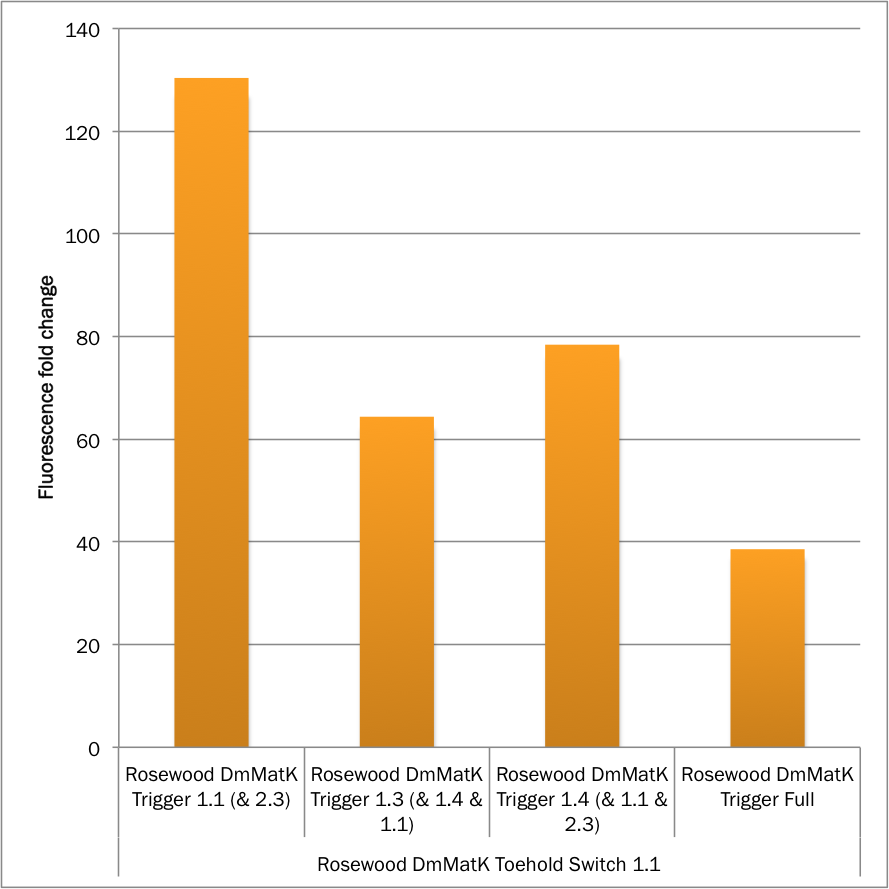
The results presented in figures 2, 3 and 4 show that the rosewood DmMatK toehold switch 1.1 is functional and behaves as expected. When tested against its cognate trigger, this toehold switch showed fluorescence with a mean comparable to that of the positive controls (Figure 2) suggesting that we leveraged the full capacity of the expression cassette. The ON/OFF ratio of this toehold switch was greater than 100 (Figure 3), on the same order of performance as some of the best switches reported in literature.
The results presented in Figure 2 show also that the sfGFP expression was readily detected in the positive controls and that the two RBS (BBa_K2675017 and BBa_K3453005) have comparable strength. As expected, no fluorescence was detected in the negative controls as either the sfGFP gene was not present or the promoter and the RBS were absent.
Figure 2. In vivo characterization of sfGFP expression by E. coli BL21 Star™(DE3) cells harbouring the rosewood DmMatK toehold switch 1.1 and the rosewood DmMatK triggers (Table 1). The negative controls have been performed with an empty pSB3T5, pSB1C3 (no trigger) and BBa_K3453103 (no promoter, no RBS) and the positive controls with BBa_K3453104 and BBa_K3453105. The data and error bars are the mean and standard deviation of at least three measurements on independent biological replicates.
Figure 3. MEFL / Particle fold changes of the rosewood DmMatK toehold switch 1.1 in the presence of the rosewood DmMatK triggers (Table 1) compared to the MEFL / Particle value in the absence of the trigger.
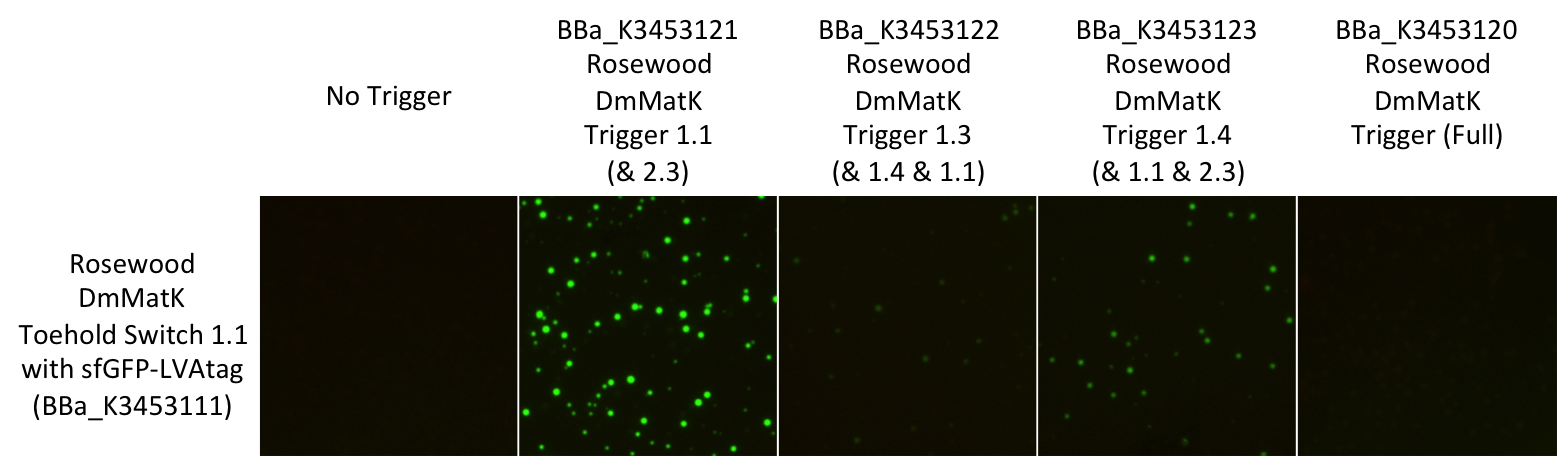
Figure 4: Pictures of E. coli BL21 Star™(DE3) cells harbouring this part in the presence of the rosewood DmMatK triggers (Table 1). The negative control was performed with an empty pSB1C3 (no trigger).
in vitro experimental characterization
An extract based cell-free system was prepared following Noireaux’s lab protocol [2] using the E. coli BL21 Star™(DE3) cells (Thermo Fisher Scientific). As our constructs are all placed under the control of T7 promoter, the T7 RNA polymerase expression was induced by adding 1 mM of IPTG to the 2YTP growth media during the production culture. Mg glutamate, K glutamate and DTT concentrations were optimized in order to have the highest efficiency of protein production achieved by the cell-free preparation. Sensor plasmids containing the toehold switch sequence in front of the sfGFP gene in pSB3T5 backbone were purified in large quantities from 300 mL LB + 10 µg/mL tetracycline cultures followed by a MaxiPrep (Macherey Nagel) plasmid extraction.
The trigger RNA was produced from a PCR reamplified template using the TranscriptAid T7 (Thermo Fisher Scientific) transcription kit. Template trigger DNA was digested using the DNAse enzyme provided in the same kit and after a RNA purification step using the Monarch® RNA Cleanup Kit (New England Biolabs) the integrity and the size of the RNA transcript was confirmed by running an aliquot of the final product on a 2.5% agarose gel. All these steps were carried in an RNAse free environment with a specific attention paid to the avoidance of external RNAse contaminations.
Each of the sensors was tested in triplicate against water and their cognate triggers. The final DNA/RNA concentrations were 5 nM and 2 µM for switches and triggers respectively. Note that the switch concentrations correspond to DNA concentrations while the trigger concentrations correspond to RNA concentrations, thus we expect similar concentrations after transcription.
Reactions were run in an opaque wall 384-well polystyrene microplate (Nunc 242764) in a final volume of 15 µL constituted by 5.33 µL cell-free extract, 6.33 µL buffer and 3.33 µL of water, DNA and RNA. The plate was incubated at 30°C and the sfGFP fluorescence (λexcitation 485 nm and λemission 528 nm) wes measured every 5 min for 8 hours in a Synergy HTX (BioTek) plate reader. An endpoint measurement was extracted at 45 minutes to compare the fluorescence of the sensor with and without addition of the trigger RNA in a fast output setup. Fluorescence values were converted into Molecules of Equivalent FLuorescein (MEFL) using the calibration curves presented on the the ‘Measurement’ page of our wiki.
When tested with its cognate trigger in the cell-free platform, BBa_K3453111 showed a positive behavior. In the presence of the trigger the fluorescence signal was strongly increased compared to the signal obtained in the absence of the trigger (Figure 5), the fluorescence fold changes reaching a significant level.
Figure 5. Characterization of sfGFP expression controlled by the rosewood DmMatK toehold switch 1.1 and the rosewood DmMatK trigger 1.1 in an E. coli BL21 Star™(DE3) based cell-free system. The data and error bars are the mean and standard deviation of at least three measurements. Fold change is defined as the ratio of the average fluorescence of sensor + trigger cell extract vs the sensor + no trigger cell extract.
References
[1] Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell (2014) 159, 925-939.
[2] Sun ZZ, Hayes CA, Shin J, Caschera F, Murray RM, Noireaux V. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. Journal of Visualized Experiments: JoVE (2013) e50762.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 149
- 1000COMPATIBLE WITH RFC[1000]