Difference between revisions of "Cell-free chassis/Commercial E. coli S30"
(→Commercial ''E. coli'' S30) |
|||
| (3 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | [[Cell-free chassis|< Back to Cell-free chassis]] | |
{| border="1" width="100%" | {| border="1" width="100%" | ||
| Line 6: | Line 6: | ||
|- | |- | ||
|<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30 <font face=georgia color=#000066 size=4>Chassis description</font>]</center> || The Commercial ''E. coli'' S30 extract was purchased from Promega. It was prepared by modifications of the method described by Zubay et al (1980). '''''E. coli'' strain B''' deficient in OmpT endoproteinase and lon protease activity was used. | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30 <font face=georgia color=#000066 size=4>Chassis description</font>]</center> || The Commercial ''E. coli'' S30 extract was purchased from Promega. It was prepared by modifications of the method described by Zubay et al (1980). '''''E. coli'' strain B''' deficient in OmpT endoproteinase and lon protease activity was used. | ||
| − | The simple constitutive gene expression device [https://parts.igem.org/wiki/index.php/Part:BBa_I13522 BBa_I13522] has been used to characterize this cell-free chassis | + | The simple constitutive gene expression device [https://parts.igem.org/wiki/index.php/Part:BBa_I13522 BBa_I13522] has been used to characterize this cell-free chassis<br> |
| + | In addition the [https://parts.igem.org/wiki/index.php/Part:BBa_F2620 BBa_F2620] device was characterised and compared to the ''in vivo'' characterisation [[Chassis/Cell-Free_Systems/Commercial_E.coli_S30/F2620| click here for comparison]] | ||
|- | |- | ||
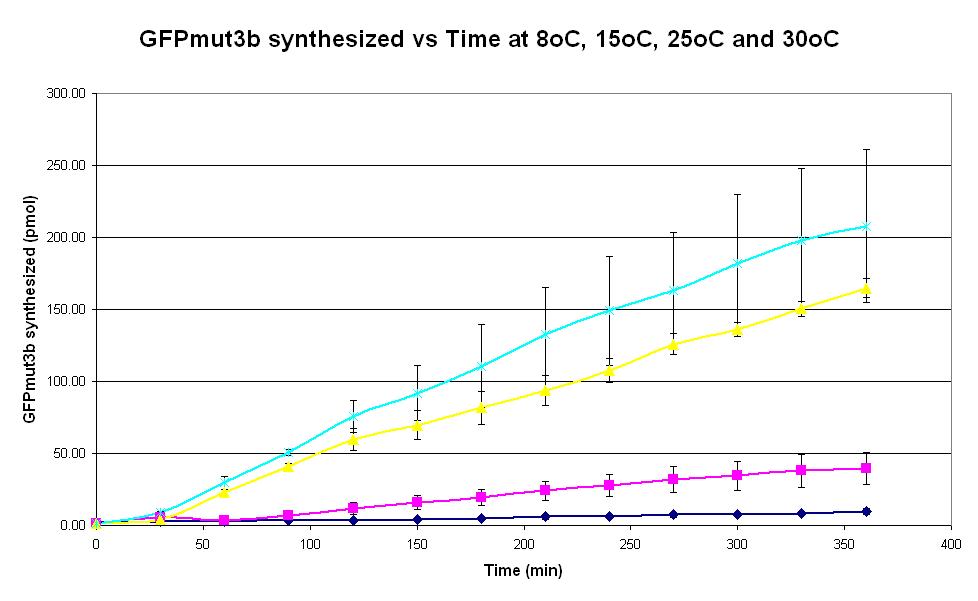
| − | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Optimum_Temp <font face=georgia color=#000066 size=4>Temperature dependence</font>]</center> || [[Image:S30_Optimum_Temp.JPG|thumb|200px|center|]] '''Graph 1. Graph of GFPmut3b synthesized (pmol) against time (minutes) at | + | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Optimum_Temp <font face=georgia color=#000066 size=4>Temperature dependence</font>]</center> || [[Image:S30_Optimum_Temp.JPG|thumb|200px|center|]] '''Graph 1. Graph of GFPmut3b synthesized (pmol) against time (minutes) at 8oC (dark blue), 15oC (pink), 20oC (yellow) and 30oC (light blue).''' The simple constitutive gene expression device [https://parts.igem.org/wiki/index.php/Part:BBa_I13522 BBa_I13522] has been used. Each of the four coloured lines show average measurements based on three replicates of the experiment. The error bars represent the standard deviation of the measurements. |
|- | |- | ||
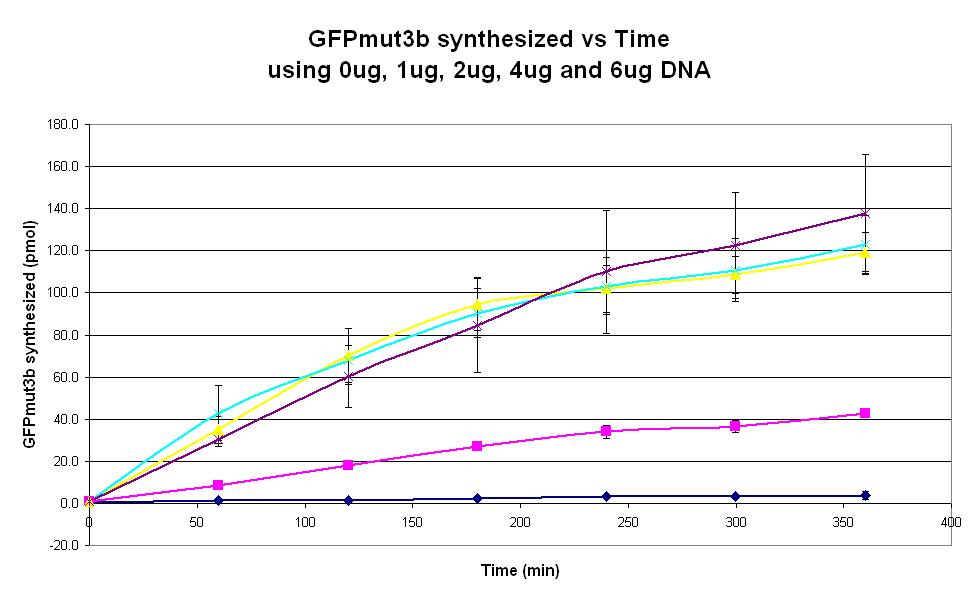
|<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Optimum_DNA <font face=georgia color=#000066 size=4>DNA quantity dependence</font>]</center> || [[Image:S30_Optimum_DNA.JPG|thumb|200px|center|]] '''Graph 2. Graph of GFPmut3b synthesized(pmol per min) against time (minutes) using 0ug (dark blue), 1ug (pink), 2ug (yellow), 4ug (light blue) and 6ug (purple).''' The simple constitutive gene expression device [https://parts.igem.org/wiki/index.php/Part:BBa_I13522 BBa_I13522] has been used. Each of the four coloured lines show average measurements based on two replicates of the experiment. The error bars represent the standard deviation of the measurements. | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Optimum_DNA <font face=georgia color=#000066 size=4>DNA quantity dependence</font>]</center> || [[Image:S30_Optimum_DNA.JPG|thumb|200px|center|]] '''Graph 2. Graph of GFPmut3b synthesized(pmol per min) against time (minutes) using 0ug (dark blue), 1ug (pink), 2ug (yellow), 4ug (light blue) and 6ug (purple).''' The simple constitutive gene expression device [https://parts.igem.org/wiki/index.php/Part:BBa_I13522 BBa_I13522] has been used. Each of the four coloured lines show average measurements based on two replicates of the experiment. The error bars represent the standard deviation of the measurements. | ||
|- | |- | ||
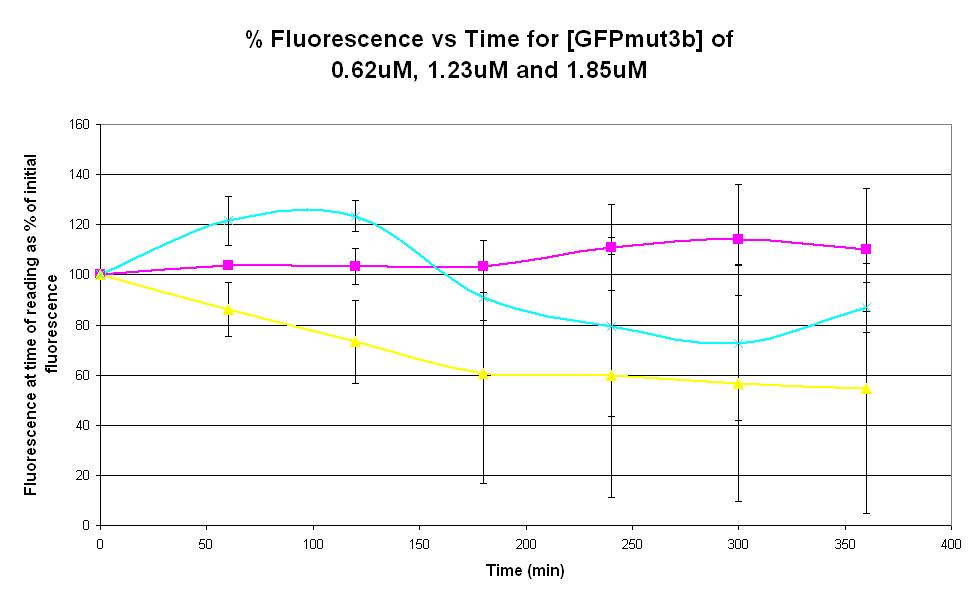
| − | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Product_Stability <font face=georgia color=#000066 size=4>Product stability</font>]</center> || [[Image:S30_Product_Stability.JPG|thumb|200px|center|]] '''Graph 3. Graph of % fluorescence against time (minutes) for [GFPmut3b] of 0.62uM (pink), 1.23uM (yellow) and 1.85uM (light blue).''' Purified GFPmut3b was used. Please click [http:// | + | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Product_Stability <font face=georgia color=#000066 size=4>Product stability</font>]</center> || [[Image:S30_Product_Stability.JPG|thumb|200px|center|]] '''Graph 3. Graph of % fluorescence against time (minutes) for [GFPmut3b] of 0.62uM (pink), 1.23uM (yellow) and 1.85uM (light blue).''' Purified GFPmut3b was used. Please click [http://2007.igem.org/Imperial/Wet_Lab/Protocols/Prot1.6 here] for the protocol for purification of GFPmut3b. Each of the three coloured lines show average measurements based on two replicates of the experiment. The error bars represent the standard deviation of the measurements. |
|- | |- | ||
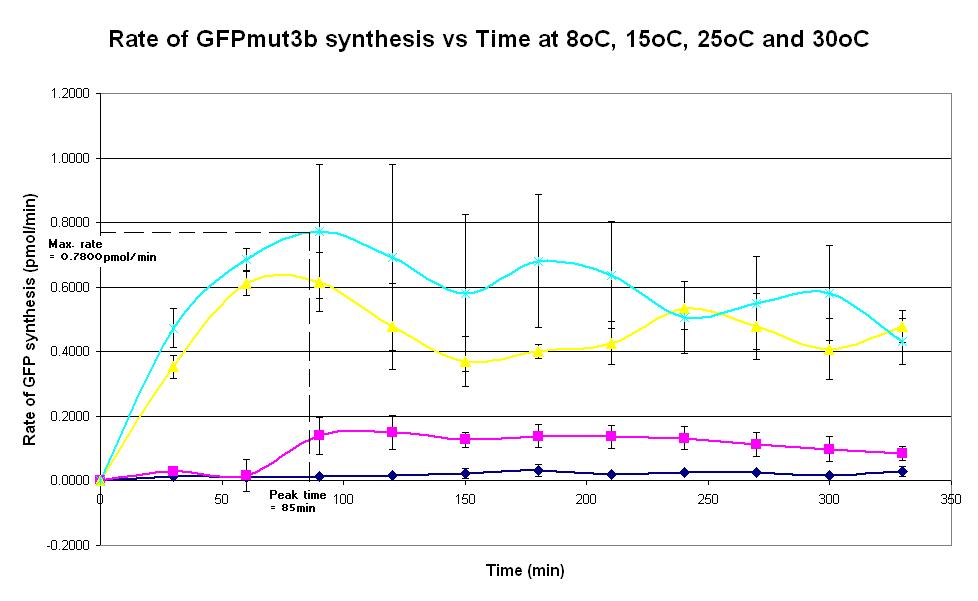
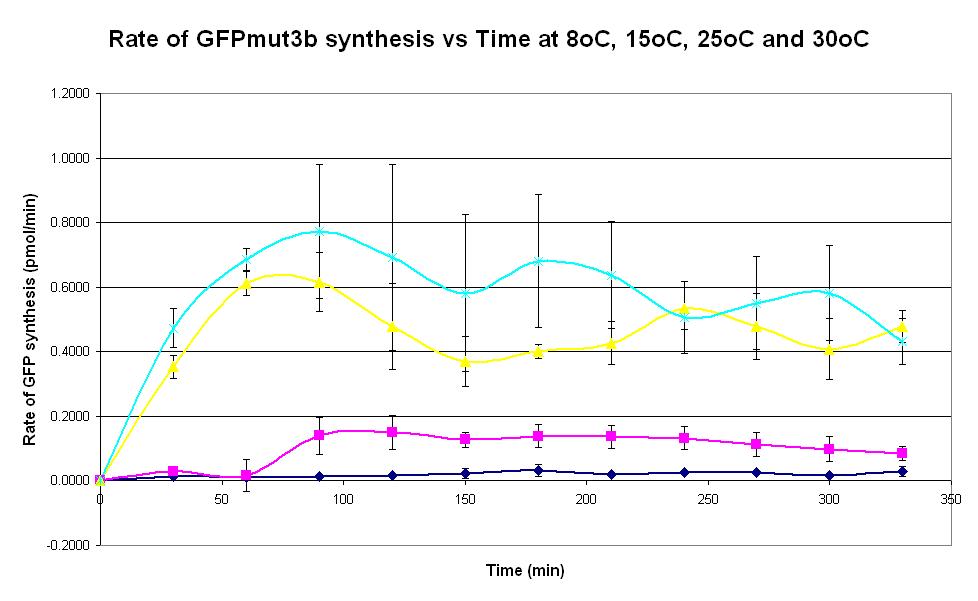
| − | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Peak_Time <font face=georgia color=#000066 size=4>Peak time</font>]</center> || [[Image:S30_Peak_Time.JPG|thumb|200px|center|]]'''Graph 4. Graph of rate of GFPmut3b synthesis (pmol per min) against time (minutes) at | + | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Peak_Time <font face=georgia color=#000066 size=4>Peak time</font>]</center> || [[Image:S30_Peak_Time.JPG|thumb|200px|center|]]'''Graph 4. Graph of rate of GFPmut3b synthesis (pmol per min) against time (minutes) at 8oC (dark blue), 15oC (pink), 20oC (yellow) and 30oC (light blue).''' The simple constitutive gene expression device [https://parts.igem.org/wiki/index.php/Part:BBa_I13522 BBa_I13522] has been used. Each of the four coloured lines show average measurements based on three replicates of the experiment. The error bars represent the standard deviation of the measurements. |
|- | |- | ||
| − | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Expression_Lifespan <font face=georgia color=#000066 size=4>Expression lifespan</font>]</center> || [[Image:S30_Expression_Lifespan.JPG|thumb|200px|center|]] '''Graph 5. Graph of rate of GFPmut3b synthesis (pmol per min) against time (minutes) at | + | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Expression_Lifespan <font face=georgia color=#000066 size=4>Expression lifespan</font>]</center> || [[Image:S30_Expression_Lifespan.JPG|thumb|200px|center|]] '''Graph 5. Graph of rate of GFPmut3b synthesis (pmol per min) against time (minutes) at 8oC (dark blue), 15oC (pink), 20oC (yellow) and 30oC (light blue).''' The simple constitutive gene expression device [https://parts.igem.org/wiki/index.php/Part:BBa_I13522 BBa_I13522] has been used. Each of the four coloured lines show average measurements based on three replicates of the experiment. The error bars represent the standard deviation of the measurements. |
|- | |- | ||
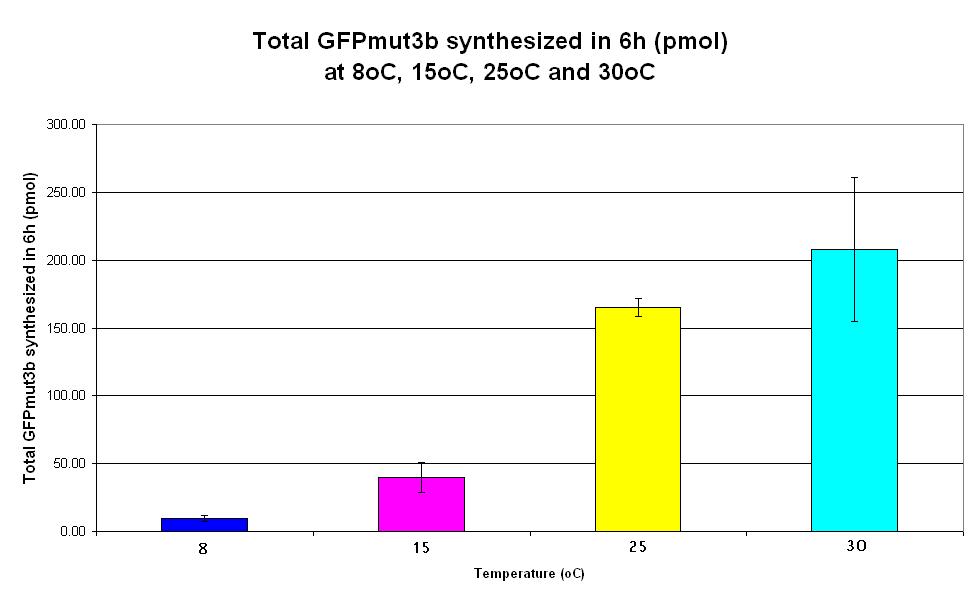
| − | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Expression_Capacity <font face=georgia color=#000066 size=4>Expression capacity</font>]</center> || [[Image:S30_Expression_Capacity.JPG|thumb|200px|center|]] '''Graph 6. Bar chart showing total GFPmut3b synthesized in 6 hours (pmol) at | + | |<center>[https://parts.igem.org/wiki/index.php/Chassis/Cell-Free_Systems/Commercial_E.coli_S30/Expression_Capacity <font face=georgia color=#000066 size=4>Expression capacity</font>]</center> || [[Image:S30_Expression_Capacity.JPG|thumb|200px|center|]] '''Graph 6. Bar chart showing total GFPmut3b synthesized in 6 hours (pmol) at 8oC (dark blue), 15oC (pink), 20oC (yellow) and 30oC (light blue).''' The simple constitutive gene expression device [https://parts.igem.org/wiki/index.php/Part:BBa_I13522 BBa_I13522] has been used. Each of the four coloured bars show average measurements based on three replicates of the experiment. The error bars represent the standard deviation of the measurements. |
|} | |} | ||
Latest revision as of 19:08, 13 February 2009
| Cell-Free Systems (back to Intro) | Introduction |
| The Commercial E. coli S30 extract was purchased from Promega. It was prepared by modifications of the method described by Zubay et al (1980). E. coli strain B deficient in OmpT endoproteinase and lon protease activity was used.
The simple constitutive gene expression device BBa_I13522 has been used to characterize this cell-free chassis | |
| Graph 1. Graph of GFPmut3b synthesized (pmol) against time (minutes) at 8oC (dark blue), 15oC (pink), 20oC (yellow) and 30oC (light blue). The simple constitutive gene expression device BBa_I13522 has been used. Each of the four coloured lines show average measurements based on three replicates of the experiment. The error bars represent the standard deviation of the measurements. | |
| Graph 2. Graph of GFPmut3b synthesized(pmol per min) against time (minutes) using 0ug (dark blue), 1ug (pink), 2ug (yellow), 4ug (light blue) and 6ug (purple). The simple constitutive gene expression device BBa_I13522 has been used. Each of the four coloured lines show average measurements based on two replicates of the experiment. The error bars represent the standard deviation of the measurements. | |
| Graph 3. Graph of % fluorescence against time (minutes) for [GFPmut3b] of 0.62uM (pink), 1.23uM (yellow) and 1.85uM (light blue). Purified GFPmut3b was used. Please click [http://2007.igem.org/Imperial/Wet_Lab/Protocols/Prot1.6 here] for the protocol for purification of GFPmut3b. Each of the three coloured lines show average measurements based on two replicates of the experiment. The error bars represent the standard deviation of the measurements. | |
| Graph 4. Graph of rate of GFPmut3b synthesis (pmol per min) against time (minutes) at 8oC (dark blue), 15oC (pink), 20oC (yellow) and 30oC (light blue). The simple constitutive gene expression device BBa_I13522 has been used. Each of the four coloured lines show average measurements based on three replicates of the experiment. The error bars represent the standard deviation of the measurements. | |
| Graph 5. Graph of rate of GFPmut3b synthesis (pmol per min) against time (minutes) at 8oC (dark blue), 15oC (pink), 20oC (yellow) and 30oC (light blue). The simple constitutive gene expression device BBa_I13522 has been used. Each of the four coloured lines show average measurements based on three replicates of the experiment. The error bars represent the standard deviation of the measurements. | |
| Graph 6. Bar chart showing total GFPmut3b synthesized in 6 hours (pmol) at 8oC (dark blue), 15oC (pink), 20oC (yellow) and 30oC (light blue). The simple constitutive gene expression device BBa_I13522 has been used. Each of the four coloured bars show average measurements based on three replicates of the experiment. The error bars represent the standard deviation of the measurements. |
References
Please refer to the Promega cell extract user guide for detailed information about the Commercial E. coli S30 extract.