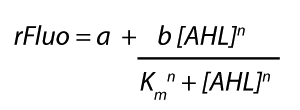Difference between revisions of "Part:BBa J10050:Experience"
| Line 24: | Line 24: | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| − | = Characterization of | + | = Characterization of weak promoter (BBa_J23113) and weak RBS (BBa_B0031) = |
| − | + | ||
| − | + | ||
We used an ''E. coli'' TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and promoter [https://parts.igem.org/Part:BBa_R0079 pLas (BBa_R0079)] followed by [https://parts.igem.org/Part:BBa_B0034 RBS (BBa_B0034)] and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts [https://parts.igem.org/Part:BBa_B0040 BBa_B0040] and [https://parts.igem.org/Part:BBa_B0015 BBa_B0015] were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and a strong promoter [https://parts.igem.org/Part:BBa_J23100 (BBa_J23100)] chosen from the [https://parts.igem.org/Promoters/Catalog/Anderson Anderson promoter collection] followed by one of the three different regulators ([https://parts.igem.org/Part:BBa_C0062 LuxR], [https://parts.igem.org/Part:BBa_C0179 LasR], and [https://parts.igem.org/Part:BBa_C0171 RhlR]) used in the experiments in order to quantify crosstalk with [https://parts.igem.org/Part:BBa_I14017 pRhl]. The detailed regulator construct design and full sequences (piG0040, piG0041, piG0042, piG0060) are [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. In the following, we describe the experimental set-up and all the different levels of crosstalk we have assessed. | We used an ''E. coli'' TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and promoter [https://parts.igem.org/Part:BBa_R0079 pLas (BBa_R0079)] followed by [https://parts.igem.org/Part:BBa_B0034 RBS (BBa_B0034)] and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts [https://parts.igem.org/Part:BBa_B0040 BBa_B0040] and [https://parts.igem.org/Part:BBa_B0015 BBa_B0015] were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and a strong promoter [https://parts.igem.org/Part:BBa_J23100 (BBa_J23100)] chosen from the [https://parts.igem.org/Promoters/Catalog/Anderson Anderson promoter collection] followed by one of the three different regulators ([https://parts.igem.org/Part:BBa_C0062 LuxR], [https://parts.igem.org/Part:BBa_C0179 LasR], and [https://parts.igem.org/Part:BBa_C0171 RhlR]) used in the experiments in order to quantify crosstalk with [https://parts.igem.org/Part:BBa_I14017 pRhl]. The detailed regulator construct design and full sequences (piG0040, piG0041, piG0042, piG0060) are [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. In the following, we describe the experimental set-up and all the different levels of crosstalk we have assessed. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
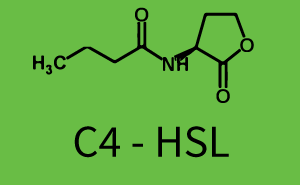
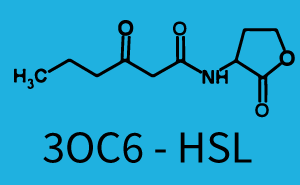
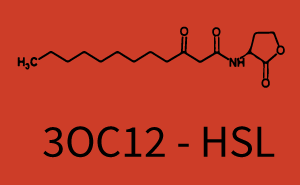
[[File:ETH Zurich 1crosstalkPrhl.png|400px|thumb|center| '''Figure 1 Overview of possible crosstalk of the [https://parts.igem.org/Part:BBa_C0171 RhlR]/[https://parts.igem.org/Part:BBa_I14017 pRhl] system with three different [[AHL|AHLs]].''' Usually, [[AHL|C4-HSL]] binds to its corresponding regulator, [https://parts.igem.org/Part:BBa_C0171 RhlR], and activates the [https://parts.igem.org/Part:BBa_I14017 pRhl] promoter (green). However, [https://parts.igem.org/Part:BBa_C0171 RhlR] may also bind [[AHL|3OC12-HSL]] (red) or [[3OC6HSL|3OC6-HSL]] (light blue) and then unintentionally activate [https://parts.igem.org/Part:BBa_I14017 pRhl].]] | [[File:ETH Zurich 1crosstalkPrhl.png|400px|thumb|center| '''Figure 1 Overview of possible crosstalk of the [https://parts.igem.org/Part:BBa_C0171 RhlR]/[https://parts.igem.org/Part:BBa_I14017 pRhl] system with three different [[AHL|AHLs]].''' Usually, [[AHL|C4-HSL]] binds to its corresponding regulator, [https://parts.igem.org/Part:BBa_C0171 RhlR], and activates the [https://parts.igem.org/Part:BBa_I14017 pRhl] promoter (green). However, [https://parts.igem.org/Part:BBa_C0171 RhlR] may also bind [[AHL|3OC12-HSL]] (red) or [[3OC6HSL|3OC6-HSL]] (light blue) and then unintentionally activate [https://parts.igem.org/Part:BBa_I14017 pRhl].]] | ||
Revision as of 11:04, 21 October 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_J10050
User Reviews
UNIQ499aec2c138489f4-partinfo-00000000-QINU
|
••••
ETH Zurich 2014 |
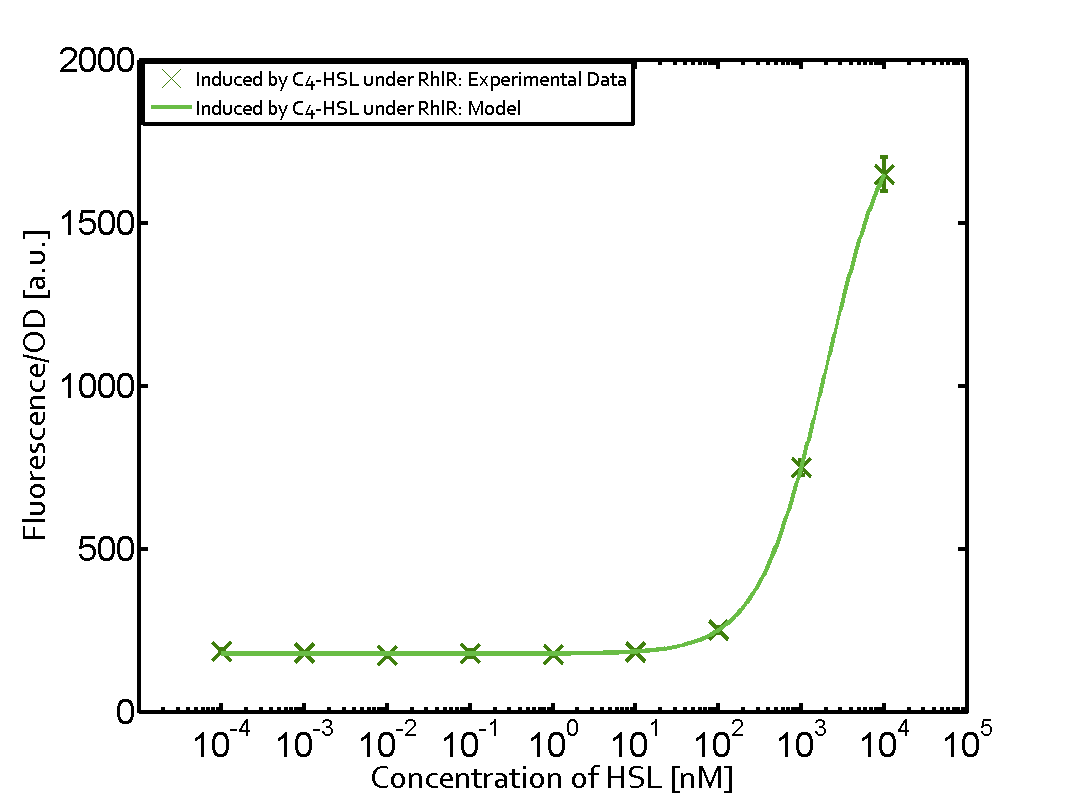
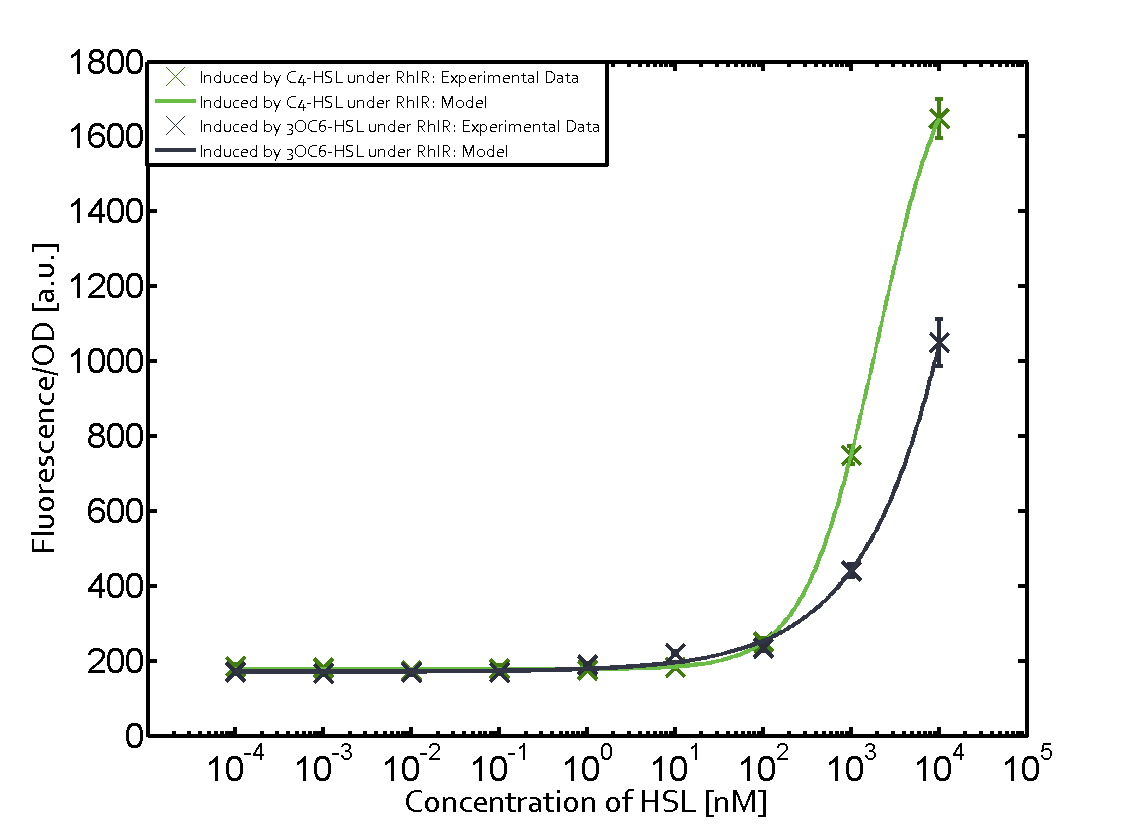
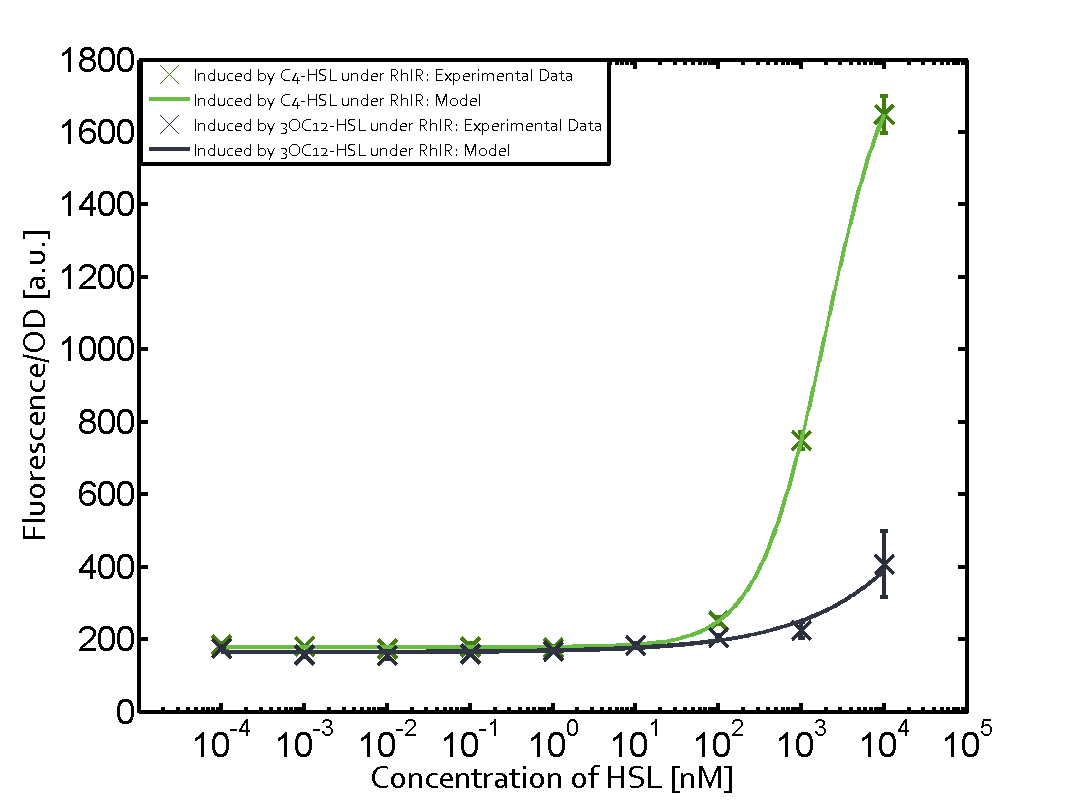
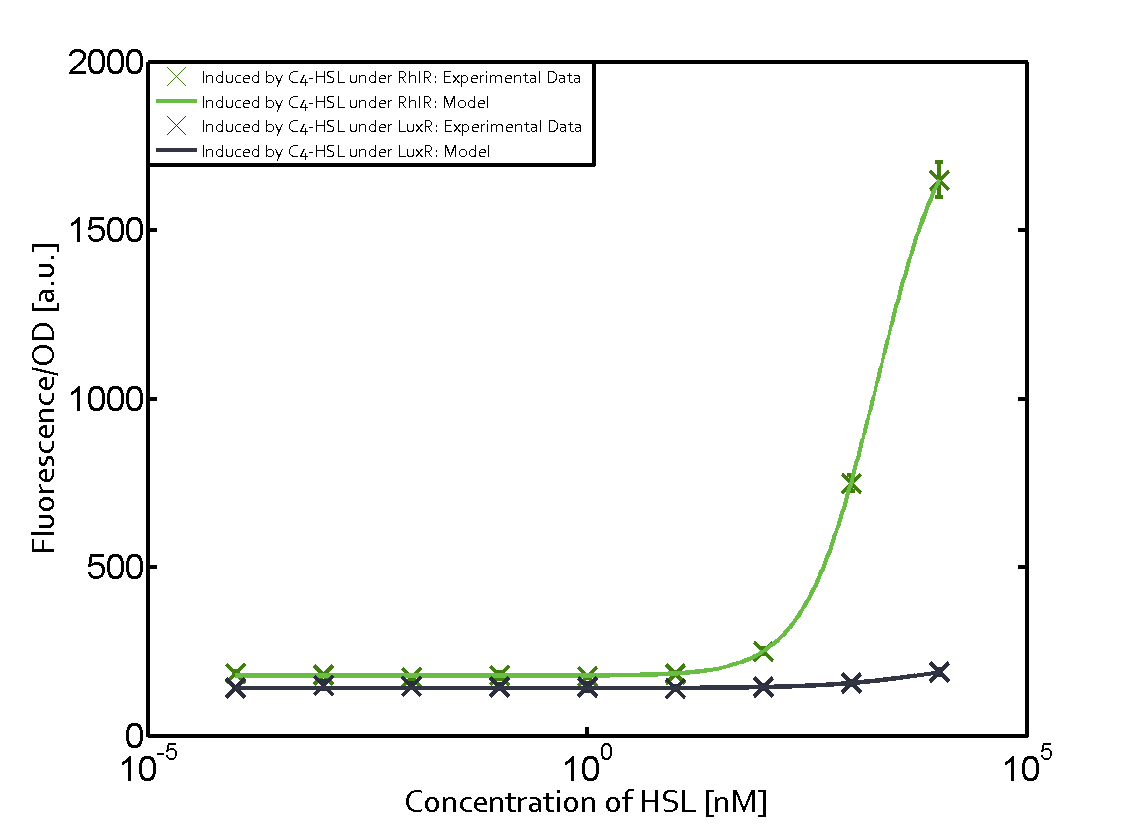
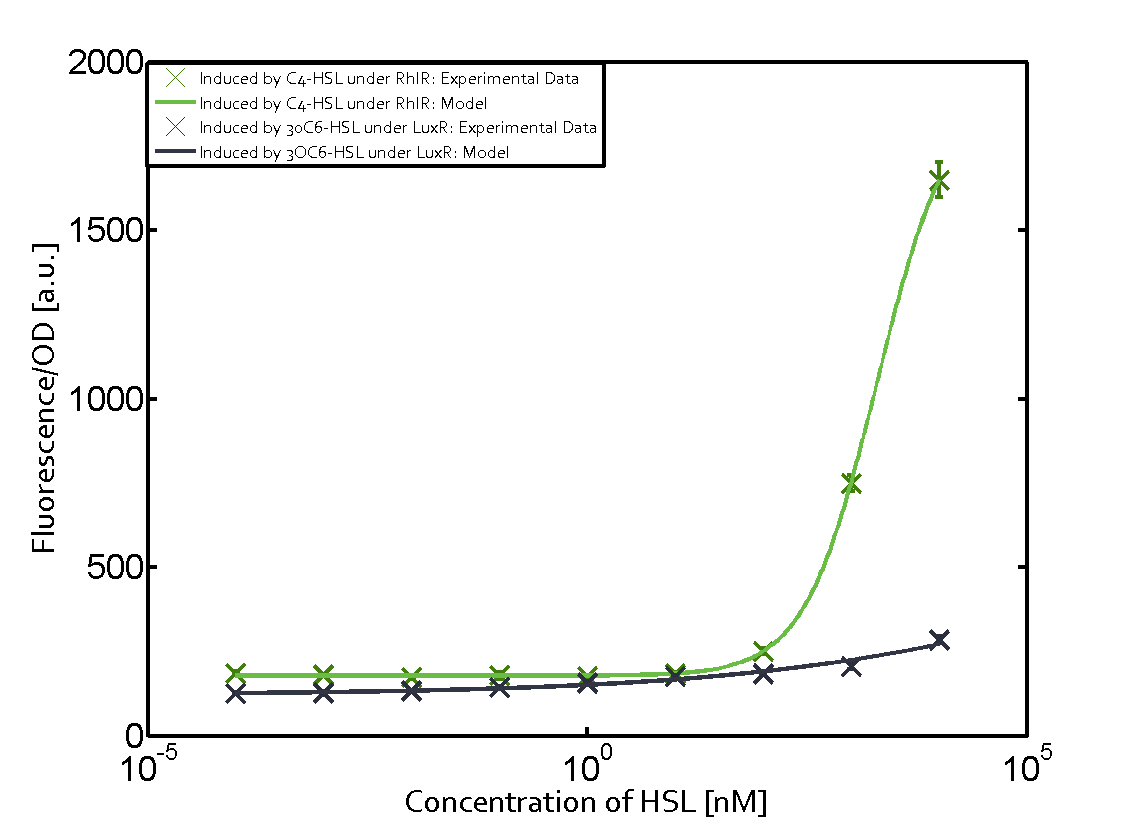
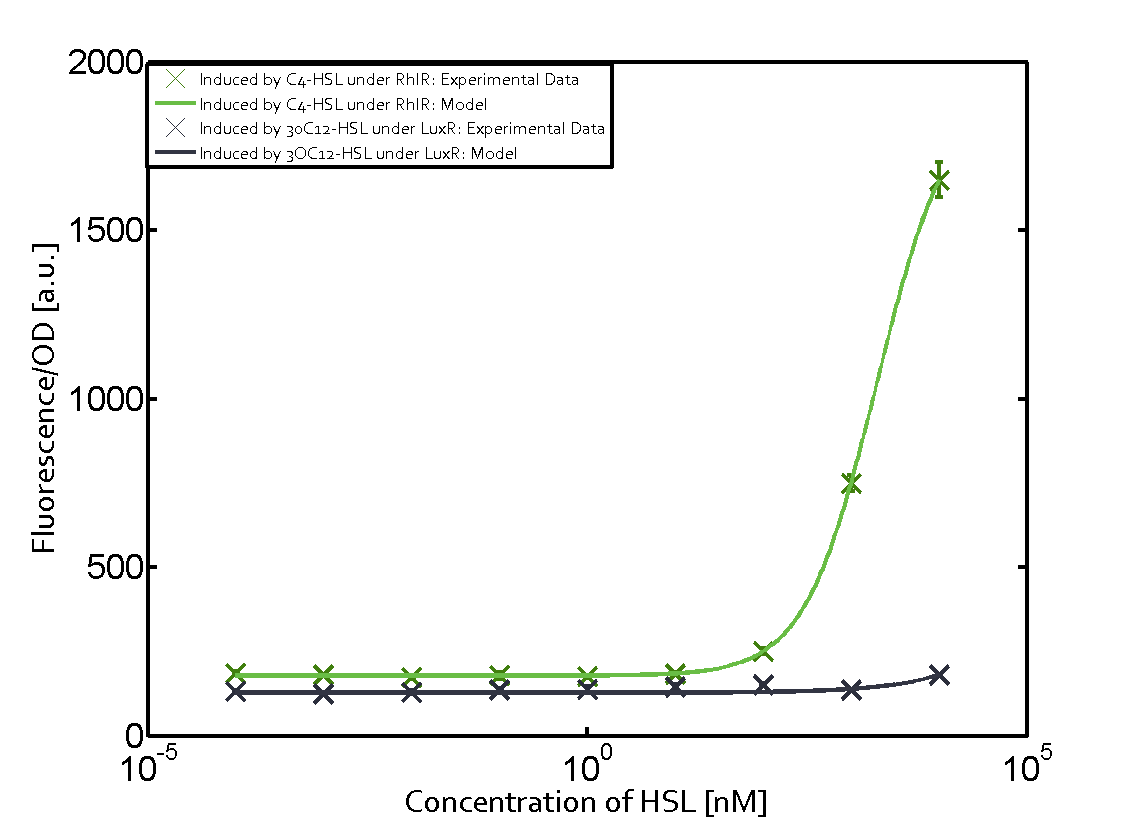
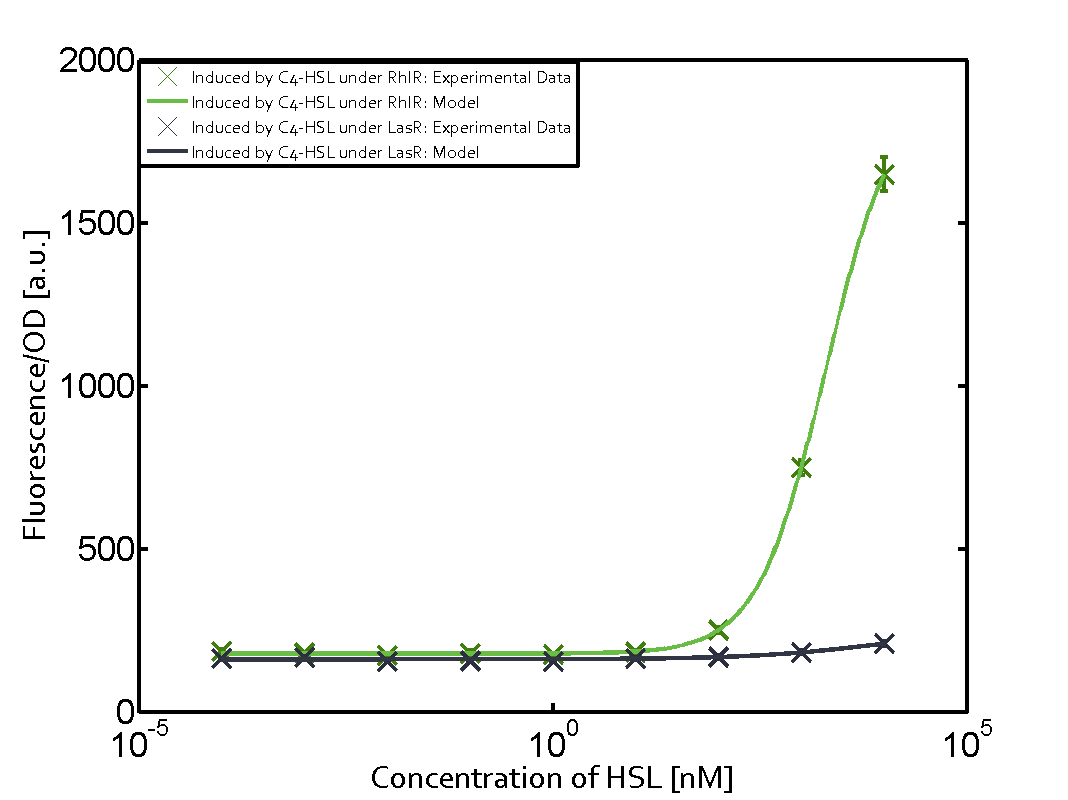
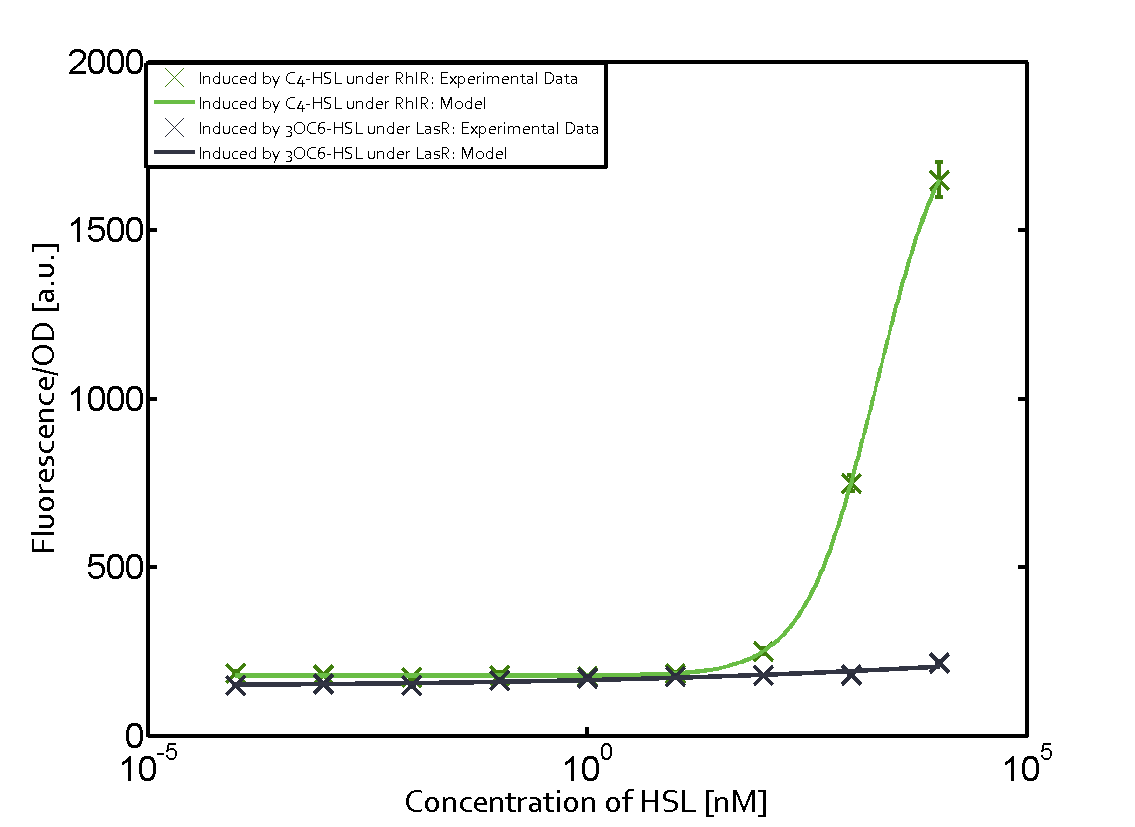
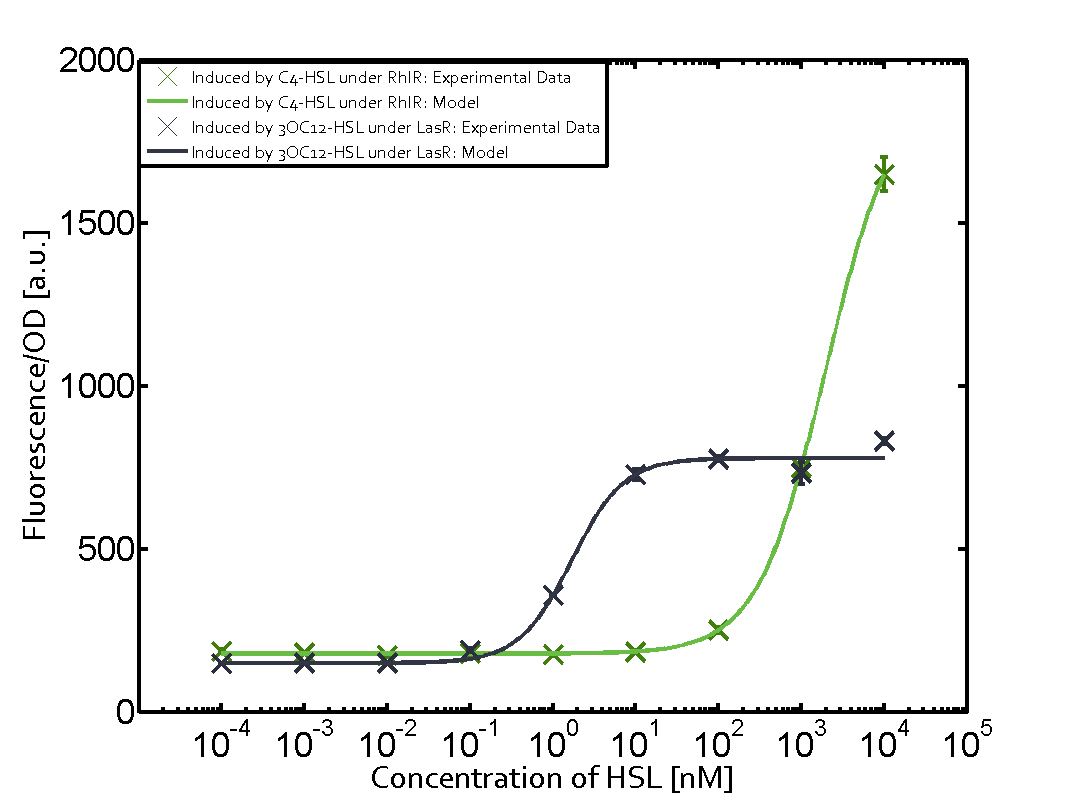
Characterization of weak promoter (BBa_J23113) and weak RBS (BBa_B0031)We used an E. coli TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and promoter pLas (BBa_R0079) followed by RBS (BBa_B0034) and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts BBa_B0040 and BBa_B0015 were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and a strong promoter (BBa_J23100) chosen from the Anderson promoter collection followed by one of the three different regulators (LuxR, LasR, and RhlR) used in the experiments in order to quantify crosstalk with pRhl. The detailed regulator construct design and full sequences (piG0040, piG0041, piG0042, piG0060) are [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. In the following, we describe the experimental set-up and all the different levels of crosstalk we have assessed. 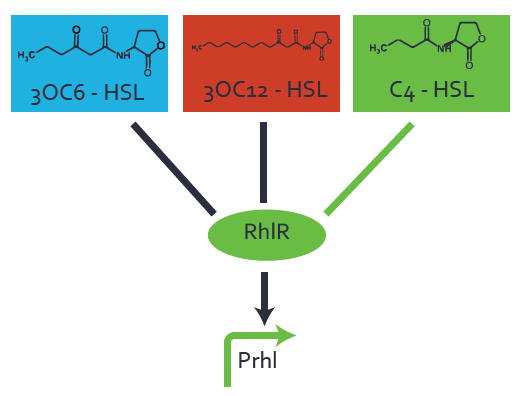 Figure 1 Overview of possible crosstalk of the RhlR/pRhl system with three different AHLs. Usually, C4-HSL binds to its corresponding regulator, RhlR, and activates the pRhl promoter (green). However, RhlR may also bind 3OC12-HSL (red) or 3OC6-HSL (light blue) and then unintentionally activate pRhl. Second Level crosstalk: other regulatory proteins, like LuxR and LasR, bind to their natural AHL substrate and activate the pRhl promoterIn the conventional system C4-HSL binds to its corresponding regulator, RhlR, and activates the pRhl promoter (Figure 2, green). However, pRhl can potentially be activate by other regulators (LuxR, LasR), binding their corresponding regulator (figure 2, 3OC6-HSL in light blue, 3OC12-HSL in red). This leads then to unwanted gene expression (crosstalk). 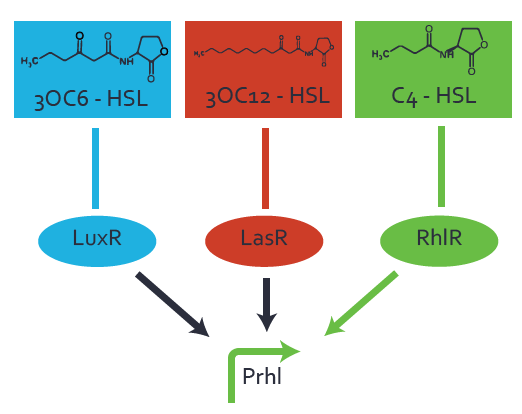 Figure 2 Overview of possible crosstalk of the RhlR/pRhl system with two additional regulators (LuxR and LasR). Usually, RhlR together with inducer C4-HSL activate their corresponding promoter pRhl (green). However, pRhl may also be activated by the LuxR regulator together with 3OC6-HSL (light blue) or by the LasR regulator together with 3OC12-HSL (red). Second order crosstalk: Combination of both cross-talk levelsThe second order crosstalk describes unintended activation of pRhl by a mixture of both the levels described above. The regulator and inducer are being different from RhlR and C4-HSL, respectively, and at the same time they do not belong to the same module. For example, the inducer 3OC6-HSL (light blue), usually binding to the regulator LuxR, could potentially interact with LasR regulator (red) and together activate pRhl (green). This kind of crosstalk is explained in Figure 3. 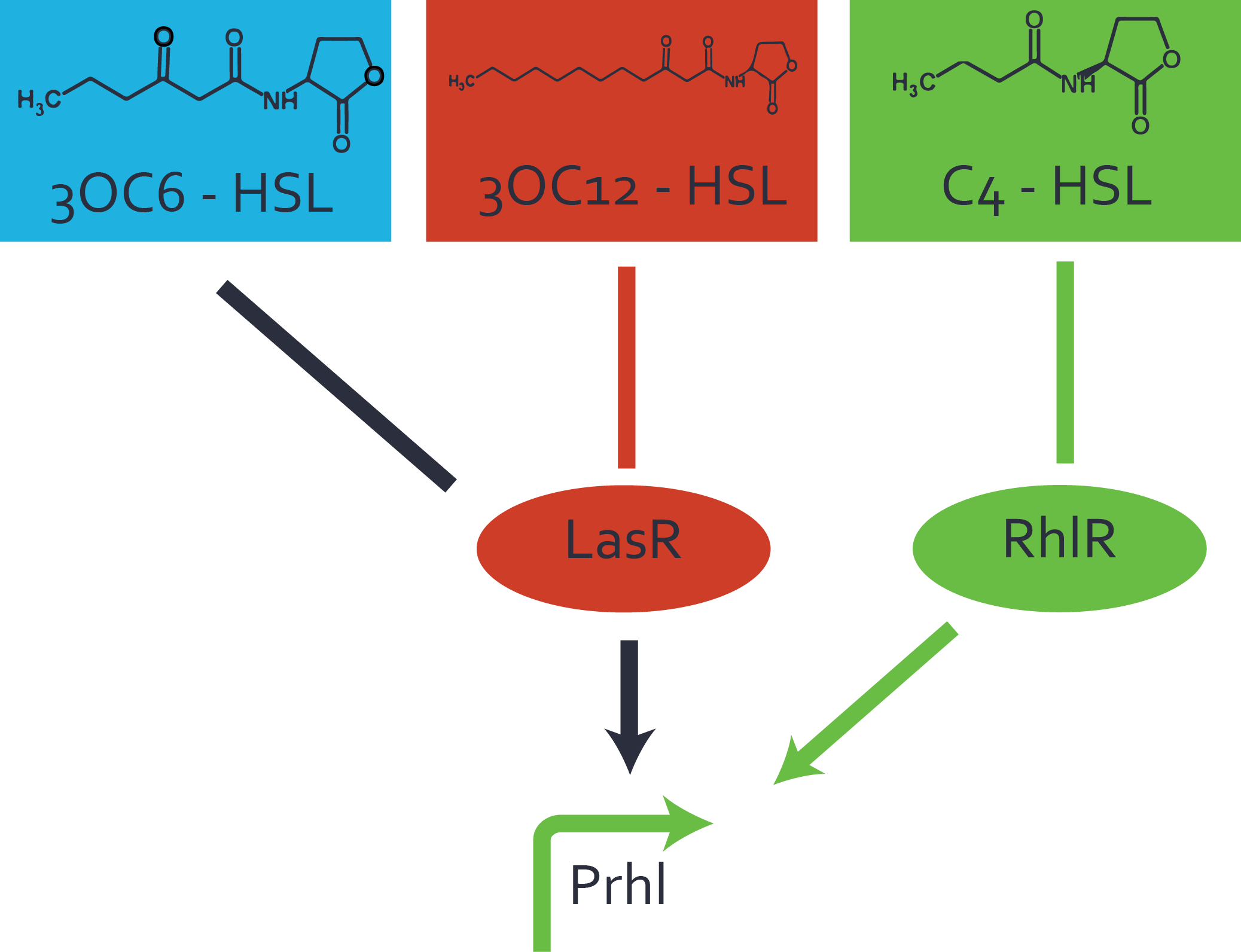 Figure 3 Overview of possible crosstalk of the pRhl promoter with both the regulator and inducer being unrelated to the promoter and each other. Usually, RhlR together with inducer C4-HSL activate their corresponding promoter pRhl (green). However, pRhl may also be activated by another regulator together with an unrelated inducer. For example, the inducer 3OC6-HSL (light blue) may interact with the LasR regulator (red) and together activate pRhl (green). Results
Modeling crosstalkEach experimental data set was fitted to an Hill function using the Least Absolute Residual method. The fitting of the graphs was performed using the following equation :
| ||||||||||||||||||||||||||||||||||||
|
Antiquity |
This review comes from the old result system and indicates that this part did not work in some test. |
UNIQ499aec2c138489f4-partinfo-00000003-QINU