Part:BBa_K1758100
Translation enhancing 5-UTR containing g10-L RBS
This sequence contains a 5' untranslated region (5'-UTR) and a strong ribosomal binding site from bacteriophage T7, named g10-L. This sequence greatly enhances translation of a following gene. The enhancing effect relies on the regulation of mRNA binding to and release of the ribosome S30 subunit (for details see Olins et al. 1989 and Takahashi et al. 2013).
g10-L is a strong RBS in E. coli but also well suited for foreign gene expression (Rangwala et al. 1991).
Usage and Biology
This part includes a RBS and a translation enhancing sequence. It improves the expression of a following gene when cloned in front of it compared to other RBS, for example B0034. The sequence of this part is derived from Olins et al. 1989 and ribosome binding analysis from Takahashi et al. 2013. When using this sequence, we recommend to clone it via Gibson assembly or similar methods, as the classical biobrick scar may have adverse effects on performance (see Lentini et al. 2013).
Design considerations

- AATAATTTTGTTTTAACTTTAA
- To increase the efficiency of translation initiation, the T7 g10 leader sequence can be employed. This sequence contains the socalled epsilon motif TTAACTTTA (Spirin and Swartz, 2008), and was originally described by Olins et al. 1988. It enhances the binding of the mRNA to the 16 S rRNA (Olins et al. 1988; Olins, Rangwala 1989; Takahashi et al. 2013), and Olins et al. showed that this sequence can massively improve heterologous gene expression in E. coli (Olins et al. 1988).
- poly-A-spacer
- With kinetic studies, Takahashi et al. showed that a spacer between the epsilon motive and the RBS improves the translation rate in vitro. This works when the spacer does not interact with the 30S subunit of the ribosome, which is the case for example for an all-adenine spacer (Takahashi et al. 2013). They further determined a 10-A-spacer as suitable when using E. coli S30 extracts instead of the PURE system.
- GAAGGAG
- The effect of the ribosome binding site (RBS) on in vitro translation has been investigated extensively (Karig et al. 2012; Lentini et al. 2013; Takahashi et al. 2013). Most important for an efficient reaction is a defined distance (4-9 bases) of the RBS to the following start codon (Karig et al. 2012; Lentini et al. 2013). The strength of the RBS has a minor effect as long as it is not too weak (Karig et al. 2012; Takahashi et al. 2013).
- AATAATCT
- According to Lentini et al. 2013, the sequence composition between RBS and start codon affects the expression level of the following gene. An AT-rich region gives the best results, whereas expression is lower with the biobrick scar TACTAGAG for example (Lentini et al. 2013)
Characterization
For characterization experiments, please see BBa_K1758101, BBa_K1758102 and BBa_K1758106. As this sequence was frequently used in our project, you can have a look at our wiki.Additional characterization by NortheasternU-Boston 2017
Additional verification was done on the function of this part in a separate cell-free expression context than previously conducted. This 5'UTR component was used with all of our parts for producing Antimicrobial peptides and other proteins. Shown below are images showing successful cell-free production via NEB PURExpress of functional Superfolder GFP, and two of our Antimicrobial peptides (AMPs) BP100 BBa_K2431002, and Micrococcin C7 BBa_K2431000. The AMPs were produced using two different T7 promoters: BBa_I712074 and BBa_I719005.
GFP expressed using NEB PURExpress kit with the transcription enhancing 5' UTR under the control of BBa_I712074. Clear expression is visible validating the basic functionality of the part.
No Template Control: D & N


Above plates show the lack of bacterial killing due to PURExpress reactions without AMP template added. "N" denotes the normal reaction concentration and "D" represents a 1:4 dilution.
Part 11: BP100 (BBa_K2431002)




The above plate based bacterial killing assay shows the effective killing of BP100 produced via PURExpress. "L" and "S" represent promoters BBa_I712074 and BBa_I719005 respectively. "N" denotes the normal reaction concentration and "D" represents a 1:4 dilution. You can read more about experimental details on NortheasternU-Boston Experiments
Part 12: Micrococcin C7 (BBa_K2431000)




The above plate based bacterial killing assay shows the effective killing of Microccon C7 produced via PURExpress. "L" and "S" represent promoters BBa_I712074 and BBa_I719005 respectively. "N" denotes the normal reaction concentration and "D" represents a 1:4 dilution. You can read more about experimental details on NortheasternU-Boston Experiments
Additional characterizazion by iGEM TU Darmstadt 2019
A similar 5'-UTR was used by iGEM TU Darmstadt 2019 to improve BBa_K1141000, thereby creating BBa_K3187014.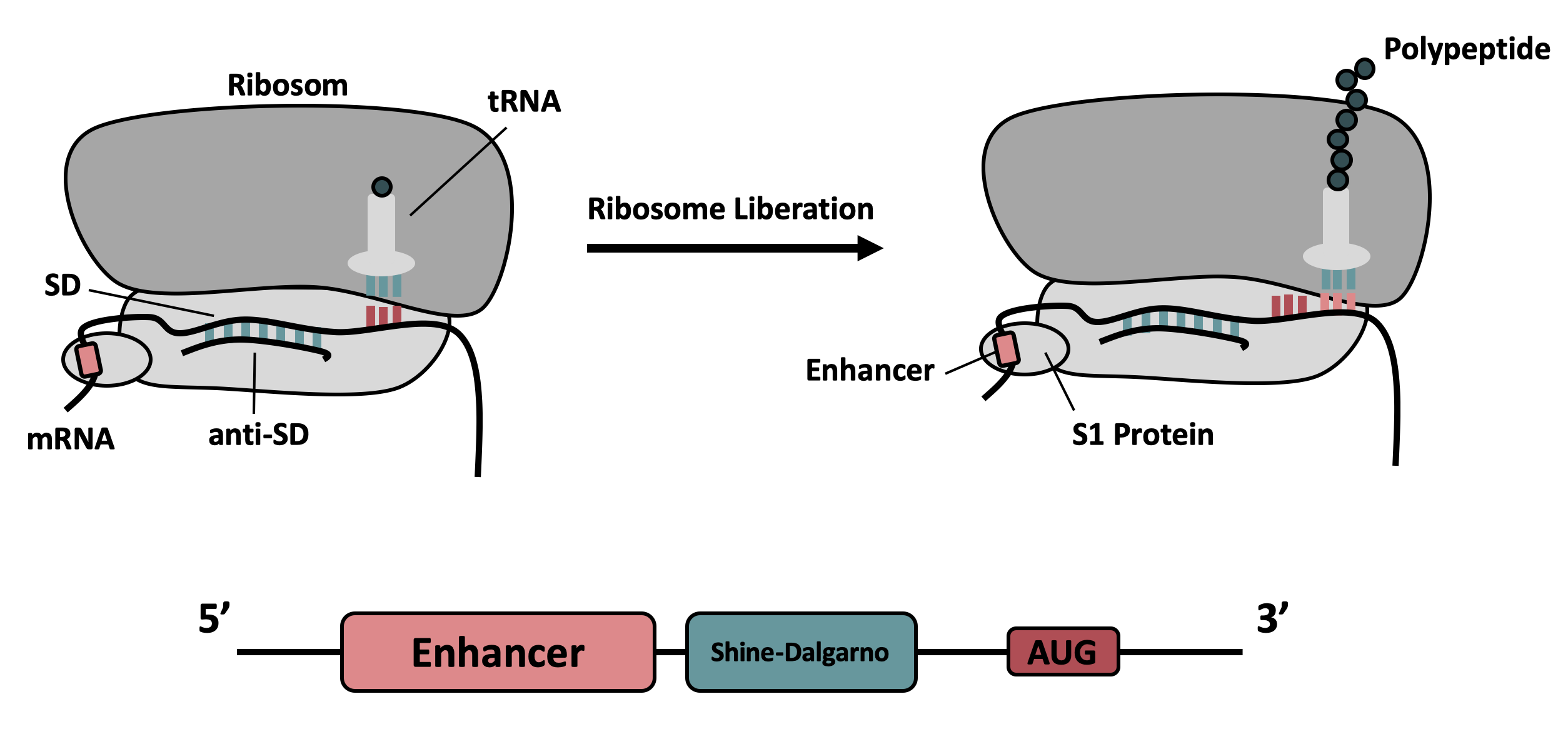
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |
