Ribosome Binding Site/Cat page/2
| RBS part table RBS part icon: |
Ribosome Binding Sites
The RBS controls the accuracy and efficiency with which the translation of mRNA begins. A ribosome binding site (RBS) is a segment of the 5' (upstream) part of an mRNA molecule that binds to the ribosome to position the message correctly for translation initiation. In bacteria, translation initiation almost always requires both an RBS sequence and a start codon. In the registry, protein coding sequences begin with the start codon. So, if you want to build a BioBrick system that produces a protein, you need to pick an RBS part and put it upstream of the protein coding sequence you want to translate.
Note that an RBS is often defined as just that part of the mRNA sequence that binds to the ribosome, however, the surrounding sequence can also affect the translation initiation rate. Consequently, a BioBrick™ RBS part contains the classic RBS but sometimes additional surrounding sequence.
This page describes the different types of RBSs that are available from the registry. While we have different classes of RBSs, most people simply want an RBS to let them express some protein of interest in bacteria. In that case, you probably need look no further than the first table below, where we list our families of high quality bacterial RBSs.
Constitutive prokaryotic RBS
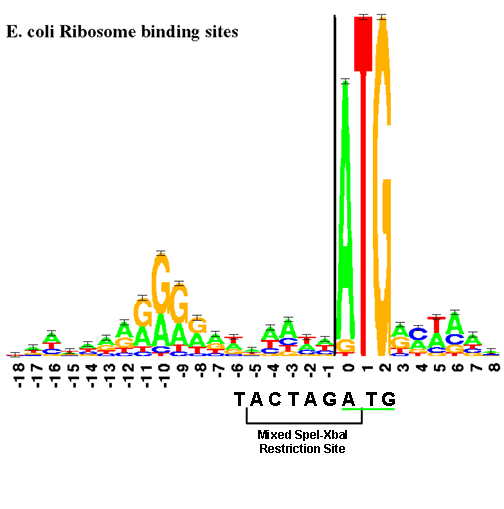
The Registry has a few collections of RBSs that are recommended for general protein expression in E. coli and other prokaryotic hosts. They are all constitutive (unregulated) RBS. Each family has multiple members covering a range of translation initiation rates. For each RBS family below, the master nucleotide sequence is shown from which members of the family are derived, an N indicates that the base location was randomized. Click on a particular family below to read more and to get sequences of specific family members.
| Family Name | Master Sequence (and bracketing sequences in grey) |
Number of Members |
|---|---|---|
| Anderson RBS library | TCTAGAGAAAGANNNGANNNTACTAGATG | 40 |
| Community Collection | TCTAGAGATTAAAGAGGAGAAATACTAGATG | 8 |
In addition to the RBS families listed above, the registry has a collection of miscellaneous constitutive prokaryotic RBSs which can be found here. Note: While we are supportive of researchers developing and contributing individual new RBSs to the Registry, we think it makes most sense if we develop a small number of high-quality RBS collections with RBS that have been used in multiple contexts and for which quantitative data exists. That way when people come along to use an RBS, they have the best chance of finding one that does what they want. For this reason, we are particularly encourage users to use RBSs from the families listed above and to contribute back data on their performance or improvements you have made to the RBSs in the collections. If you'd like to undertake constructing a high-quality new and interesting RBS family that's great too!
Regulated Prokaryotic RBS (Riboregulators)
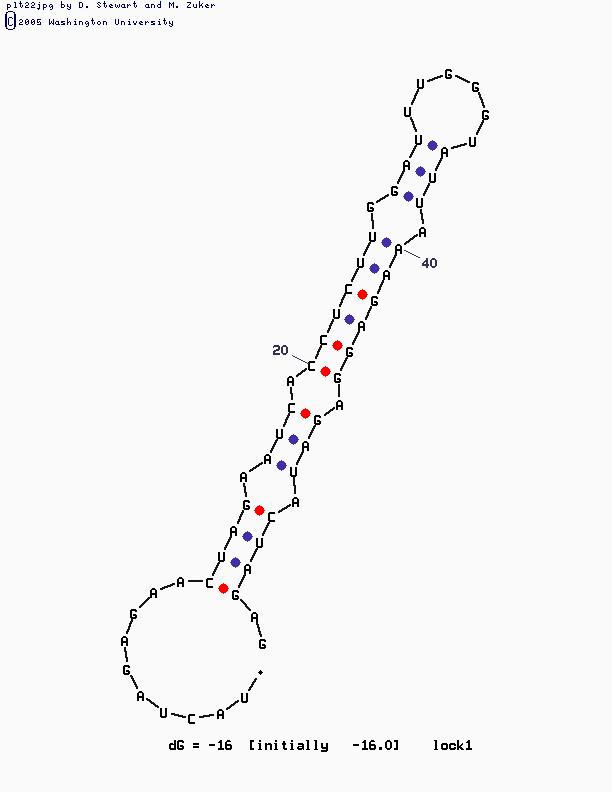
A regulated RBS is one for which the binding affinity of the RBS and the ribosome can be controlled, thereby changing the RBS strength. One strategy for regulating the strength of prokaryotic RBSs is to control the accessibility of the RBS to the ribosome. By occluding the RBS in RNA secondary structure, translation initiation can be significantly reduced. By contrast, by reducing secondary structure and revealing the RBS, translation initiation rate can be increased. Isaacs and coworkers Isaacs engineered mRNA sequences with an upstream sequence partially complementary to the RBS. Base-pairing between the upstream sequence and the RBS 'locks' the RBS off. A 'key' RNA molecule that disrupts the mRNA secondary structure by preferentially base-pairing with the upstream sequence can be used to expose the RBS and increase translation initiation rate. The Registry has a small collection of prokaryotic RBSs that can be regulated in this manner that were developed by the Berkeley iGEM team in 2005 building on the work of Isaacs and coworkers.
| Family Name | Master Sequence (and bracketing sequences in grey) |
Number of Members |
|---|---|---|
| Isaacs RBS library | TCTAGAGAACTAGAATCACCTCTTGGATTTGGGTATTAAAGAGGAGATACTAGATG | 2 |
Yeast RBS
Yeast RBSs, more often known as Kozak sequences, are designed to be recognized by the yeast ribosome. As a eukaryotic translation signal, the sequence of yeast RBSs are distinct from prokaryotic RBSs. The registry collection of yeast RBSs is currently small, we need your help! Please contribute new yeast RBSs to the registry or provide further information about our existing yeast RBSs.
| Name | Sequence | Description | Relative Strength | Predicted Strength |
|---|---|---|---|---|
| BBa_J63003 | cccgccgccaccatggag | designed yeast Kozak sequence | n/a | |
| BBa_K165002 | cccgccgccaccatggag | Kozak sequence (yeast RBS) | ||
| BBa_K792001 | ggatccacgattaaaagaatg | Kozak sequence from yeast α-factor mating pheromone (MFα1) |
Constitutive Custom RBS
Prokaryotic ribosomes recognize RBSs primarily via base-pairing between the RBS and an unstructured end of the 16s rRNA molecule that forms part of the ribosome. Ribosomes can be designed with mutations in the portion of the 16s rRNA molecule that binds to the RBS. These mutant ribosomes can be used to translate mRNA that aren't recognized by wild-type ribosomes. This mechanism can be used to tightly regulate the translation rate of particular mRNA. These mRNA must have mutant RBS that have a sequence very different from the Shine-Dalgarno sequence. The Registry has a collection of these RBS based on the work of Oliver Rackham and Jason Chin Rackham and developed by Barry Canton.
| Family Name | Master Sequence (and bracketing sequences in grey) |
Number of Members |
|---|---|---|
| Rackham RBS library | TCTAGAGCACCACTACTAGATG | 3 |
References
<biblio>
- Isaacs pmid=15208640
</biblio>