Part:BBa_R0011
Promoter (lacI regulated, lambda pL hybrid)
Inverting regulatory region controlled by LacI (BBa_C0010, BBa_C0012, etc.) The PLlac 0-1 promoter is a hybrid regulatory region consisting of the promoter P(L) of phage lambda with the cI binding sites replaced with lacO1. The hybrid design allows for strong promotion that can nevertheless be:
- repressed by LacI, the Lac inhibitor (i.e. repressor) (BBa_C0012) ([LUTZ97]).
- induced by [http://openwetware.org/wiki/IPTG IPTG] in E.Coli DH5-alpha-Z1 (same paper reference) over a >600-fold range
Intrinsic Noise Value: 0.0040 (compare with R0010: 0.0707; R0051: 0.0869). See [http://2015.igem.org/Team:William_and_Mary William_and_Mary iGEM 2015]
>Internal Priming Screening Characterization of BBa_R0011: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Usage and Biology
Strong promoter. [jb, 5/24/04]
R0011 will be on in strains without lacI, off in strains that are lacIq (ie. Part:BBa_V1003) and medium in strains that are lacI
2019 ZJU-China’s Characterization
The Optimal IPTG Concentration and Temperature for the Expression of Protein in E.coli BL21 (DE3) with the PLIac01 Hybrid Promoter (BBa_R0011) and pSB1C3 and the Relation between the Strength of BBa_R0011 and IPTG Concentration at the Transcription Level
I Overview
In this year, we constructed an EGFP expression vector (with pSB1C3 as the plasmid skeleton) in which the expression of EGFP was controlled by the PLIac01 Hybrid Promoter (BBa_R0011) and transformed it into E.coli BL21 (DE3) which is commonly used to express protein. Then, by measuring the EGFP fluorescence, we investigate the optimal IPTG concentration and temperature for the expression of protein in E.coli BL21 (DE3) using the BBa_R0011 in pSB1C3. In the measurement, the E.coli BL21 (DE3) which contained plasmid only consisting of EGFP and pSB1C3 was used as the blank. Meanwhile, we carried out qRT-PCR to directly investigate the relation between the original strength of BBa_R0011 and IPTG concentration at the transcription level without interfering by context.
II Results
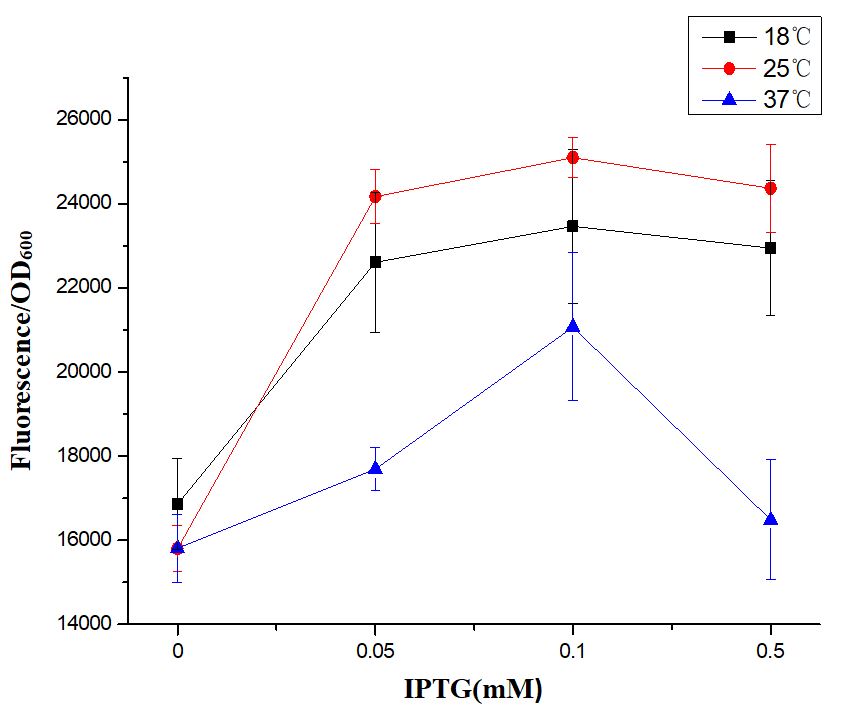
Fluorescence Characterization Shows that the Optimal Protein Expression Condition: 0.1mM IPTG & 25℃
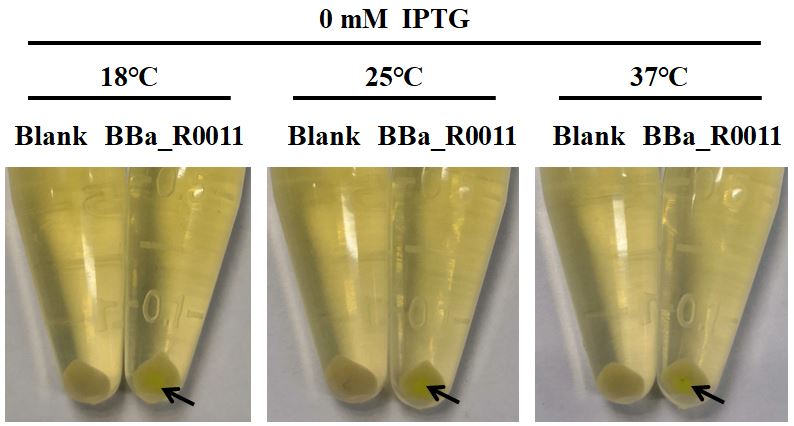
First of all, we found that when the IPTG concentration was 0 mM (no inducer was added), the expression of EGFP was visible to the naked eye (Fig. 1), indicating that when BBa_R0011 was used to express protein in E.coli BL21 (DE3) with pSB1C3 as the plasmid skeleton, it is “leaky”. We measured the EGFP green fluorescence of E.coli BL21 (DE3) when the concentration of IPTG was 0 mM, 0.05 mM, 0.1 mM and 0.5 mM at 18℃, 25℃ and 37℃, respectively. The results showed that the optimal condition for using BBa_R0011 to express protein in the given background was: 0.1 mM IPTG & 25℃ culture temperature. For IPTG concentration, no matter which culture temperature, the green fluorescence intensity increased with the increase of IPTG concentration and reached the maximum value when the IPTG concentration was 0.1mM, and then it decreased with the further increase of IPTG concentration. We speculated that this is because the IPTG greater than 0.1mM inhibited the growth of E.coli BL21 (DE3), therefore, the expression of EGFP decreased. For the culture temperature, except for the condition where IPTG concentration is 0 mM, the green fluorescence intensity at 25℃ is always higher than that at other two culture temperatures, which may be because the 25℃ is conducive to the growth of E.coli BL21 (DE3) compared with 18℃, and is conducive to the expression of foreign genes compared with 37℃.
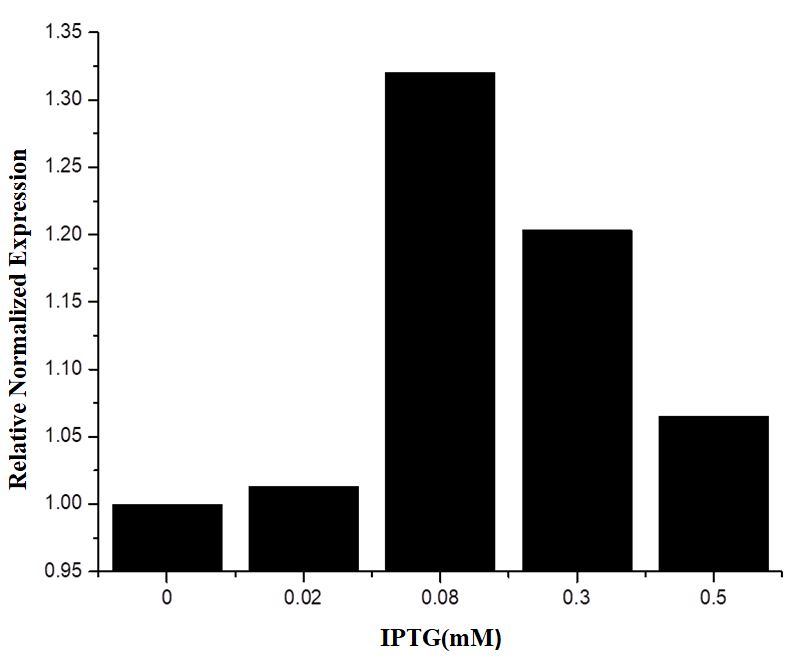
The Relation between the Strength of BBa_R0011 and IPTG Concentration at the Transcription Level
From the results of fluorescence characterization, 25℃ seems to be a more favorable temperature for the opening of BBa_R0011 in p SB1C3 in E.coli BL21 (DE3), so we chose this temperature as the culture temperature for qRT-PCR. We selected two frequently used housekeeping genes RecA and 16S rRNA as reference genes to compare the relative expression level of EGFP controlled by BBa_0011 under different IPTG concentrations of 0 mM, 0.02 mM, 0.08 Mm, 0.3 mM and 0.5 mM at 25℃. At IPTG concentration between 0 and 0.08 mM, the strength of BBa_0011 increases significantly with the increase of IPTG concentration. The relative normalized expression of EGFP at the IPTG concentration of 0.08 mM is 1.32 times that at the IPTG concentration of 0 mM, which is the highest value in 5 test groups. After the IPTG concentration exceeds 0.08 mM, the strength of BBa_0011 gradually decreases with the increase of IPTG concentration.
III Protocol
Fluorescence Characterization
- Construct the expression vector of EGFP initiated by BBa_0011 and the vector containing only EGFP, both with pSB1C3 as the plasmid skeleton.
- Transform these two kinds of plasmids into E. coli BL21(DE3) to obtain “blank” and “BBa_R0011”.
- Pick a single colony from “blank” and “BBa_R0011”, respectively, and inoculate in 5mL liquid LB medium + 5uL Chloramphenicol (25mg/mL in EtOH) and grow the cells overnight at 37 °C and 220 rpm.
- Inoculate 50uL of each overnight culture into 4.95 mL LB with Chloramphenicol to make 12 parallel groups of “blank” and “BBa_R0011”, respectively, and grow the cells at 37 °C and 220 rpm until OD600 reaches 0.6.
- Add IPTG to final concentrations of 0(parallel 1-3), 0.05(parallel 4-6), 0.1(parallel 7-9), 0.5mM(parallel 10-12) and grow the cells at temperatures of 18℃(parallel 1,4,7,10), 25℃(parallel 2,5,8,11) and 37℃(parallel 3,6,9,12), 220 rpm for 6 hours. The IPTG concentration and culture temperature of “blank” and “BBa_R0011” with the same number are the same.
- Read and record the fluorescence intensity (excitation wavelength: 485 nm; detection wavelength: 528 nm) and OD600.
- Repeat each group for 3 times.
qRT-PCR
- Pick a single colony from “BBa_R0011” and inoculate in 5mL liquid LB medium + 5uL Chloramphenicol (25mg/mL in EtOH) and grow the cells overnight at 37 °C and 220 rpm.
- Inoculate 50uL of the overnight culture into 4.95 mL LB with Chloramphenicol to make 7 parallel groups and grow the cells at 37 °C and 220 rpm until OD600 reaches 0.6.
- Add IPTG to final concentrations of 0(parallel 1), 0.02(parallel 2), 0.05(parallel 3), 0.08(parallel 4), 0.1(parallel 5), 0.2(parallel 6), 0.5mM(parallel 7) and grow the cells at temperature of 25℃ and 220 rpm for 6 hours.
- Extracted RNA, make reverse transcription and carry out qRT-PCR with recA and 16S rRNA as reference genes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
//direction/forward
//chassis/prokaryote/ecoli
//promoter
//regulation/negative
//classic/regulatory/uncategorized
| negative_regulators | 1 |
| positive_regulators |

 1 Registry Star
1 Registry Star