Part:BBa_K3453172
Truncated Grapevine VvTrnL-UAA Toehold Switch 1.1 with sfGFP-LVAtag
This part is an sfGFP-LVAtag (BBa_K2675006) expression cassette under the control of a Truncated Grapevine Toehold Switch 1.1 lacking the trigger binding site (BBa K3453072) for sequence-based detection of Vitis vinifera.
The truncation in BBa K3453072 compared to the grapevine switch 1.1 (BBa_K2916050) concerns the trigger binding site which is absent. Thus this part mimics the open (unfolded) state of this toehold switch.
This part is a control for the functional characterisation of BBa_K2916068, BBa K3453170 and BBa K3453171.
Usage and Biology
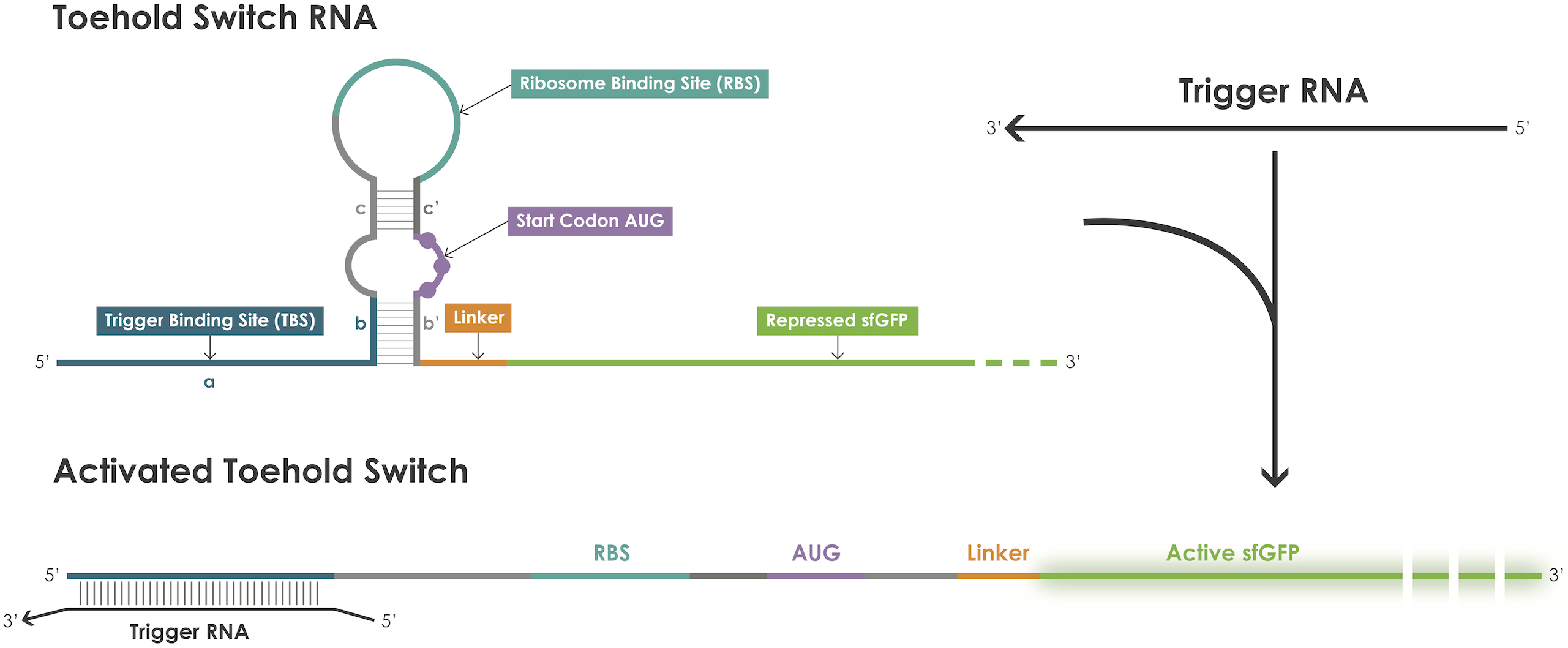
As depicted in Figure 1, a toehold switch blocks translation initiation because the RBS and the ATG start codon are embedded in the middle of a hairpin structure maintained by base pairs before and after the start codon [1]. The hairpin can be unfolded upon binding of a trigger RNA thereby exposing the RBS and the ATG start codon and thus permitting translation of the reporter protein. The binding between the trigger and the switch is dictated by RNA-RNA interactions between complementary base pairs.
Figure 1. Toehold switches principle.
In this composite part sfGFP-LVAtag (BBa_K2675006) was equipped by the Truncated Grapevine Toehold Switch 1.1 lacking the trigger binding site (BBa K3453072) and placed under the control of the T7 promoter (BBa_K2150031) and of the strong SBa_000587 synthetic terminator (BBa_K3453000). The truncation in BBa K3453072 compared to the grapevine switch 1.1 (BBa_K2916050) concerns the trigger binding site which is absent. Thus this part mimics the open (unfolded) state of this toehold switch. In BBa_K2916050 (which is the T7 promoter together with grapevine switch 1.1), the trigger binding region is in the same direction as in the grapevine genome and of the trigger sequences derived from (BBa_K3453081 and BBa_K3453083). As a result, this trigger / switch pair does not interact and is not functional. However, the hairpin structure of the grapevine switch 1.1 is predicted to be unfolded by the reverse complements of the two triggers BBa_K3453082 and BBa_K3453084 (Table 1).
| Table 1. Grapevine triggers. | |||
|---|---|---|---|
| Label | Part number | Transcriptional units’ part numbers | Source |
| Grapevine trigger 'short' | BBa_K3453081 | BBa_K3453181 | EPFL 2019 iGEM team personal communication |
| Grapevine trigger 'short reverse' | BBa_K3453082 | BBa_K3453182 | Reverse complement of BBa_K3453081 |
| Grapevine trigger 'long' | BBa_K3453083 | BBa_K3453183 | designed by the EPFL 2019 iGEM team and its sequence was deduced from “Amplification” page of the EPFL 2019 wiki where is described how the triggers were designed: the grapevine trigger was PCR amplified using as template the Vitis vinifera TrnL-UAA and the forward primers EC_fwd1 or EC_fwd1_T7 and the EC_rev1 reverse primer (it should be noted that the sequences of all reverse primers listed in the additional materials of this page are given as 3’-5’ sequences and not in the conventional 5’-3’ orientation). |
| Grapevine trigger 'long reverse' | BBa_K3453084 | BBa_K3453184 | Reverse complement of BBa_K3453083 |
BBa_K3453172 was assembled by Golden Gate in the low copy plasmid pSB3T5. For this, a fragments containing the BBa_K3453072 was synthesized flanked by BbsI type IIS restriction sites, and then inserted into BBa_K3453103, a platform in which a Golden Gate adapter with BbsI sites is present upstream of the sfGFP-LVAtag.
The trigger sequences (Table 1) were placed under the control of the T7 promoter and followed by the strong SBa_000587 synthetic terminator for the T7 RNA polymerase and the resulting transcriptional units were synthesized and assembled in the high copy plasmid pSB1C3.
For toehold switch characterisation we decided to use E. coli BL21 Star™(DE3) cells (Thermo Fisher Scientific), genotype F- ompT hsdSB (rB-mB-) gal dcm rne131 (DE3). As all BL21(DE3) E. coli strains, these cells contain the T7 RNA polymerase under control of the lacUV5 promoter and thus require IPTG to induce expression. The particularity of BL21 Star™(DE3) cells is that they contain a truncated version of the RNaseE gene (rne131) that leads to reduced level of mRNA degradation and thus increased RNA stability. For fluorescence measurements, E. coli cells containing switch and trigger plasmids were first grown overnight in 96-deep-well plates containing 1 mL of LB medium supplemented with 5 µg/mL tetracycline and 17.5 µg/mL chloramphenicol, then diluted by 40x into similar media. Upon reaching early log-phase, cells were further diluted 20x in 100 µL of LB medium supplemented with 5 µg/mL tetracycline, 17.5 µg/mL chloramphenicol and 10 µM IPTG in an opaque wall 96-well polystyrene microplate, the COSTAR 96 (Corning). The plate was incubated at 37°C at 200 rpm and the sfGFP fluorescence (λexcitation 483 nm and λemission 530 nm) and OD600nm were measured every 10 min for 24 hours in a CLARIOstar (BMGLabtech) plate reader. Fluorescence values were normalised by OD600nm and, using the calibration curves presented on the ‘Measurement’ page of our wiki, we converted the arbitrary units into Molecules of Equivalent FLuorescein (MEFL) / particle.
The results presented in figures 2 and 3 show that this part is functional: sfGFP expression was observed in the presence and in the absence of grapevine RNA, which is expected as the truncated grapevine switch 1.1 contains only an RBS and a few codons in frame with the sfGFR-LVAtag gene. Surprisingly, the sfGFP expression was low. This suggests that the sequence upstream of the natural ATG of sfGFP has a negative influence on its expression.
The results presented in Figure 2 show also that the sfGFP expression was readily detected in the positive controls and that the two RBS (BBa_K2675017 and BBa_K3453005) have comparable strength. As expected, no fluorescence was detected in the negative controls as either the sfGFP gene was not present or the promoter and the RBS were absent.
Figure 2. In vivo characterization of sfGFP expression by E. coli BL21 Star™(DE3) cells harbouring the truncated grapevine toehold switch 1.1 in BBa_K3453172 and the 'short' and 'long' grapevine triggers in both forward and reverse orientation (BBa_K3453181, BBa_K3453182, BBa_K3453183 and BBa_K3453184). The negative controls have been performed with an empty pSB3T5, pSB1C3 (no trigger) and BBa_K3453103 (no promoter, no RBS) and the positive controls with BBa_K3453104 and BBa_K3453105. The data and error bars are the mean and standard deviation of at least three measurements on independent biological replicates.
Figure 3: Pictures of E. coli BL21 Star™(DE3) cells harbouring this part and in the presence of the 'short' and 'long' grapevine triggers in both forward and reverse orientation (BBa_K3453181, BBa_K3453182, BBa_K3453183 and BBa_K3453184). The negative control was performed with an empty pSB1C3 (no trigger).
As mentioned above, the results presented in Figure 2 suggests that the sequence upstream of the natural ATG of sfGFP has a negative influence on its expression. The same sequence is present in all BBa_K2916050 (T7 promoter + grapevine switch 1.1) derived parts that we analysed (BBa_K2916068, BBa K3453170, BBa K3453171 and BBa K3453172). Indeed, translation initiation rates (Figure 4) predicted using the Salis lab RBS Calculator v2.1 [2–5] show a lower rate for BBa K3453172 compared to both positive controls BBa_K3453104 and BBa_K3453105.
Figure 4. Translation initiation rates predicted with the Salis' RBS Calculator 2.1 [2–5].
The different features of BBa_K2916050 may be responsible for this behaviour and among them the most obvious are the mRNA secondary structure and the RBS strength. To test this hypothesis, we designed two derivatives of the grapevine toehold switch 1.1 denoted 1.1.2 and 1.1.3, theirs truncated versions laking the trigger binging sites, as well as their cognate sfGFP or sfGFP-LVAtag expression cassettes.
These parts are listed in Table 2 and the results of their characterisation summarised in Figure 5. All details are available on their individual pages in the registry and the full story can be read on the 'Contribution' page of our wiki.
| Table 1. Grapevine toehold switches 1.1 and derivatives. | |||
|---|---|---|---|
| Label | Toehold Switch Part Number | Toehold Switch + sfGFP Part Number | Toehold Switch + sfGFP-LVAtag Part Number |
| Grapevine VvTrnL-UAA Toehold Switch 1.1 | BBa_K2916050 | BBa_K2916068 & BBa_K3453170 | BBa_K3453171 |
| Truncated Grapevine VvTrnL-UAA Switch 1.1 (no trigger binding site) | BBa_K3453072 | BBa_K3453172 | |
| Grapevine VvTrnL-UAA Toehold Switch 1.1.2 | BBa_K3453073 | BBa_K3453173 | BBa_K3453174 |
| Truncated Grapevine VvTrnL-UAA Switch 1.1.2 (no trigger binding site) | BBa_K3453075 | BBa_K3453175 | |
| Grapevine VvTrnL-UAA Toehold Switch 1.1.3 | BBa_K3453076 | BBa_K3453176 | BBa_K3453177 |
| Truncated Grapevine VvTrnL-UAA Switch 1.1.3 (no trigger binding site) | BBa_K3453078 | BBa_K3453178 | |
Figure 4. In vivo characterization of sfGFP expression by E. coli BL21 Star™(DE3) cells harbouring the grapevine toehold switches 1.1 (Table 2) and the 'short' and 'long' grapevine triggers in both forward and reverse orientation (BBa_K3453181, BBa_K3453182, BBa_K3453183 and BBa_K3453184). The negative controls have been performed with an empty pSB3T5, pSB1C3 (no trigger) and BBa_K3453103 (no promoter, no RBS) and the positive controls with BBa_K3453104 and BBa_K3453105. The data and error bars are the mean and standard deviation of at least three measurements on independent biological replicates.
References
[1] Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell (2014) 159: 925-939.
[2] Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nature Biotechnology (2009) 27: 946–950.
[3] Espah Borujeni A, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Research (2014) 42: 2646–2659.
[4] Espah Borujeni A, Salis HM. Translation initiation is controlled by RNA folding kinetics via a ribosome drafting mechanism. Journal of the American Chemical Society (2016) 138: 7016–7023.
[5] Espah Borujeni A, Cetnar D, Farasat I, Smith A, Lundgren N, Salis HM. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Research (2017) 45: 5437–5448.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 113
- 1000COMPATIBLE WITH RFC[1000]
| None |