Part:BBa_K1761005
LargeBit Split Luciferase
LargeBit is the big part of a Split Luciferase. Its molecular weight is 18 kDa. This construct on its own has no function since LargeBit on itself has no luminescence activity. For luminescence activity, the SmallBit of the Split Luciferace is needed.
Sequence has been published by iGEM TU-Eindhoven 2017.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Split luciferase can be used as signaling components. This split luciferase consists of two parts, namely LargeBit and SmallBit. These parts were both inserted in a pETDuet-1 vector together with OmpX and a linker. LargeBit and SmallBit have an affinity towards each other, so when in close proximity they will come together and will give a luminescence signal. Followed is a short description of each part and of the NanoBit construct.
OmpX - LargeBit BBa_K1761005 [1]
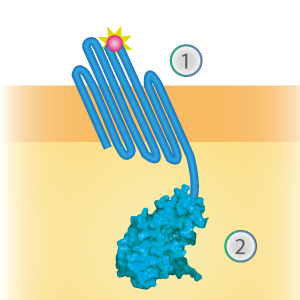
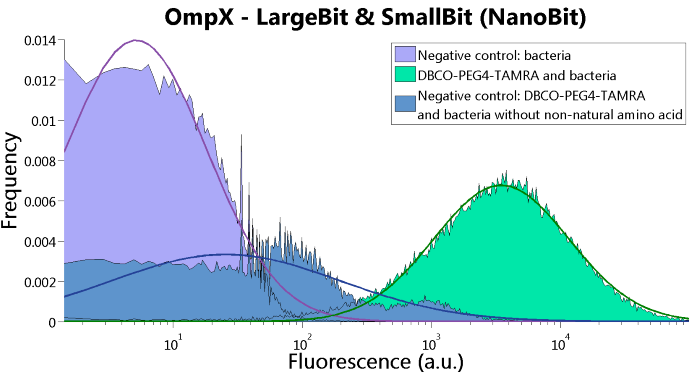
LargeBit is the big part of this Split Luciferase. Its molecular weight is 18 kDa. OmpX (1) (with a correct mutation for the amber stop codon TAG), a BamHI-linker and LargeBit (2) together will look like Figure 1. This construct on its own has no function since LargeBit on itself has no luminescence activity.
Figure 1: Schematical overview of the OmpX - LargeBit construct.
OmpX - SmallBit BBa_K1761006
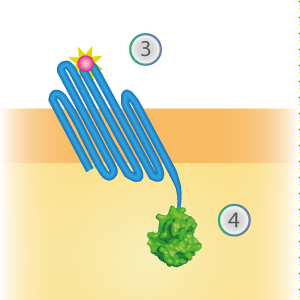
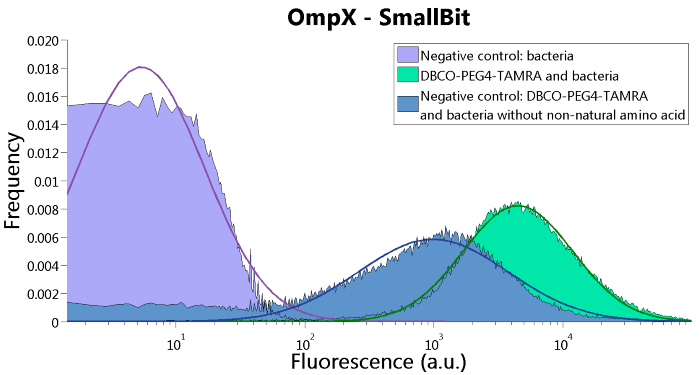
SmallBit is the small part of this Split Luciferase. Its molecular weight is 1 kDa, it is only 11 amino acids long. OmpX (1) (with a correct mutation for the amber stop codon TAG), a BsoBI-linker and SmallBit (2) together will look like Figure 2. This construct on its own has no function since SmallBit on itself has no luminescence activity.
Figure 2: Schematical overview of the OmpX - SmallBit construct.
NanoBit construct
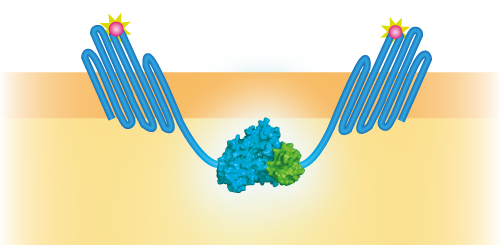
NanoBit utilizes a structural complementation-based approach to monitor protein interactions within living cells. Protein interaction promotes structural complementation and generation of a bright, luminescent enzyme. Protein dynamics can be followed in real-time in living cells following addition of the Nano-Glo Live Cell Reagent, a non-lytic detection reagent containing the cell-permeable furamizine substrate. See Figure 3 for the whole construct.
Figure 3: Schematical overview of the NanoBit construct.
Characterization
When mutated with the amber stop codon TAG, a unnatural amino acid with an azide-functionalized group can be expressed. After the expression of this amino acid, OmpX can covalently bind almost anything, as long as it contains a DBCO-functionalized group. The binding finds place by using a bio-orthogonal “click” reaction (SPAAC chemistry). To test the functionality of this “click” reaction, some experiments were done by clicking DBCO-PEG4-TAMRA at the surface. For all the experiments, the following vectors were used: pETDuet-1 with one or two construct(s) inserted (OmpX + intracellular protein) and pEVOL-pAzF (tRNA + tRNA synthetase, see BBa_K1492002 [2]). Both vectors were transformed into BL21(DE3). The expression was introduced by adding arabinose, IPTG and the unnatural amino acid.
DBCO-PEG4-TAMRA Confirmation
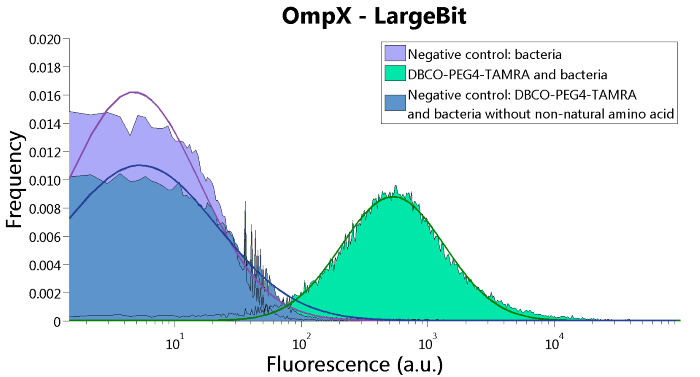
To confirm whether OmpX is in the membrane and whether or not the unnatural amino acid is being incorporated into OmpX, DBCO-PEG4-TAMRA was used. TAMRA is a fluorescent dye that can be used to verify the “click” reaction. If the unnatural amino acid is present, DBCO-PEG4-TAMRA should “click” to the transmembrane protein OmpX and stay there. This can be analyzed with FACS. For more information about how to perform FACS experiments, see our Protocol Page [http://2015.igem.org/Team:TU_Eindhoven/Project/Protocols].
To verify that OmpX is in the membrane, we used the OmpX – LgBiT and OmpX – SmBiT constructs. These gave the following results after clicking with DBCO-PEG4-TAMRA (see Figure 4, 5 and 6). From this it can be concluded that OmpX is in the membrane and that the “click” reaction works.
Figure 4: FACS results of OmpX - LargeBit.
Figure 5: FACS results of OmpX - SmallBit
Figure 6: FACS results of OmpX - LargeBit and OmpX - SmallBit.
Bioluminescence Confirmation
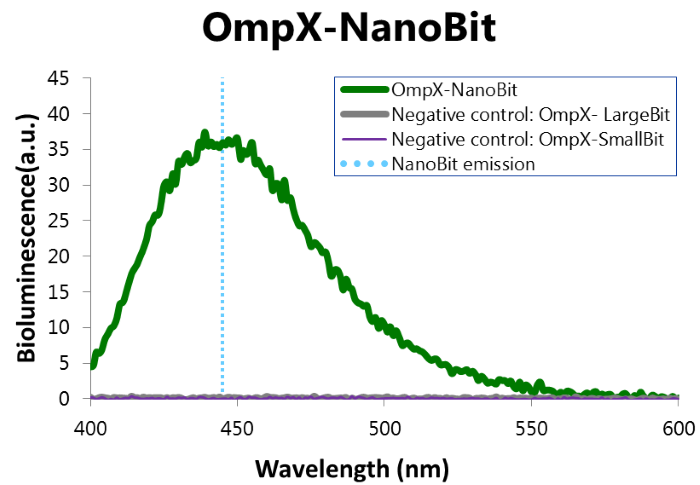
To confirm whether NanoBit is present and is working, a bioluminescence measurement was performed. The results of this experiment are shown in Figure 7.
Figure 7: Bioluminescence of results of the NanoBit construct.
Characterization iGEM 2016
Besides linking SmallBiT and LargeBiT to OmpX, it is also possible to use the NanoBiT system in other systems. iGEM TU Eindhoven 2016 (http://2016.igem.org/Team:TU-Eindhoven) used the NanoBiT system to do in vitro assays on T14-3-3 scaffold proteins. SmallBiT and LargeBiT were linked to CT52, a protein which has high affinity for the T14-3-3 scaffold if the small molecule fusicoccin is present.
CT52-SmallBiT: "https://parts.igem.org/Part:BBa_K2065000"
CT52LargeBiT: "https://parts.igem.org/Part:BBa_K2065007"
This system was used as a read out method for T14-3-3 heterodimers. Under influence of a small molecule, SmallBit and LargeBit attached to CT52 dimerize on the scaffold, leading to activation of NanoLuc Luciferase and emission of bioluminescence light. It is schematically visualised in figure 1. For more detailed information about the application of CT52-SmallBiT visit our wiki "http://2016.igem.org/Team:TU-Eindhoven/Read-out".
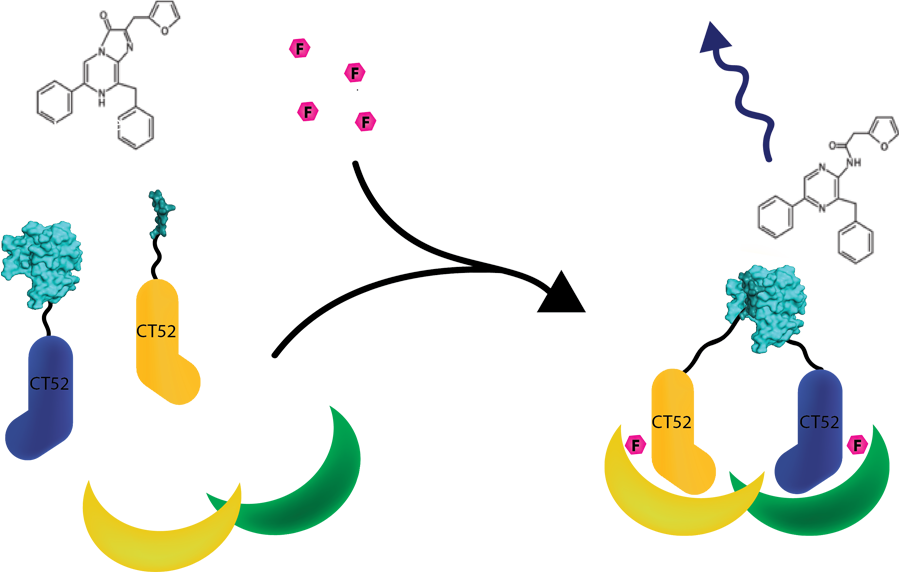
Figure 1: Measurement system for T14-3-3 heterodimers using the NanoBiT system linked to CT52. Under the influence of fusicoccin, the split NanoBits attached to CT52 will bind to T14-3-3 and emit a blue fluorescence light when dimerized and when furimazine is present.
Bioluminescence confirmation
To determine whether the split luciferase system is working, a bioluminescence signal should be visible around 460nm when SmallBiT and LargeBiT dimerize. When Furimazine is added to the dimerized Nanoluc luciferase, the Furimazine is converted into Furimamide. When this reaction occurs, light with a wavelength of 460 nm will be emitted. The spectrum of the CT52-NanoBiT system is visualed below.
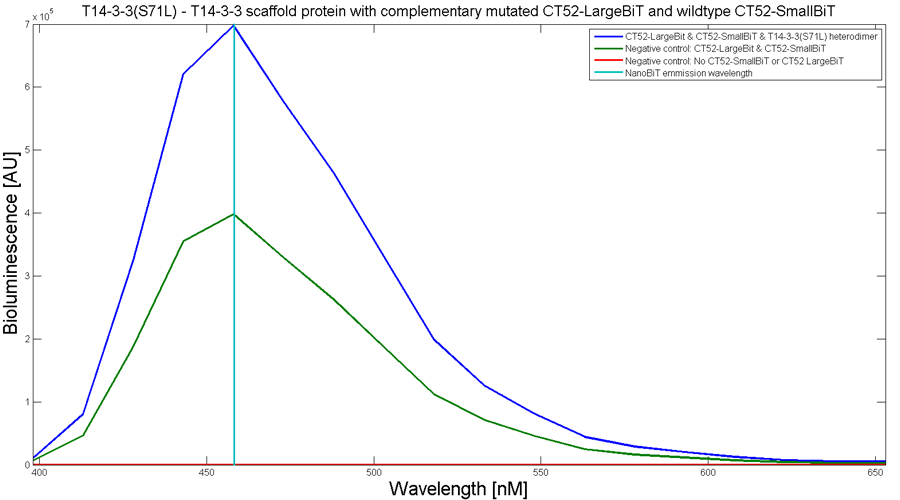
Figure 2: Bioluminescence spectrum with CT52(I947H)-LargeBiT and CT52-SmallBiT, assembled on T14-3-3(S71L) - T14-3-3
An example of a measurement which can be done on a heterodimer is a T14-3-3 heterodimer concentration gradient assay, in which the functionality can be verified. This measurement is shown below for T14-3-3(S71L) - T14-3-3. More measurements on T14-3-3 heterodimer scaffold with the CT52-NanoBiT system can be found on our wiki "http://2016.igem.org/Team:TU-Eindhoven/Results".
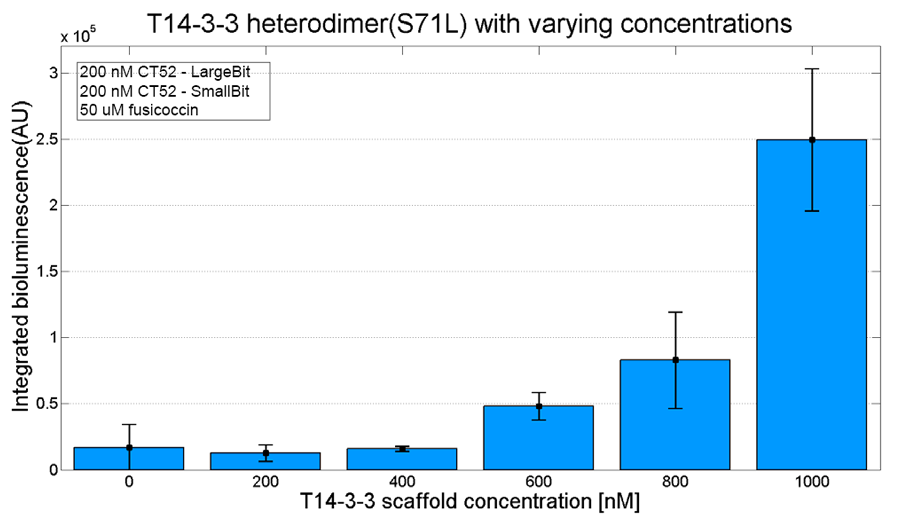
Figure 3: T14-3-3 functionality measurement with varying scaffold concentrations from 0 nM to 1 µM T14-3-3(S71L/I72V)-T14-3-3. 50 µM fusicoccin was used for all measurements. Mutant forms CT52 (I947F)-LargeBiT and CT52(wildtype)-SmallBiT were used with a concentration of 200 nM. The substrate used was 1250x diluted from stock.
NanoBiT calibration
In order to analyse the performance of our CT52 fused NanoBiT fragments we measured the luminescence they produce at varying concentrations from 10nM to 10mM.
After global optimization using metropolis Monte Carlo and Latin hypercube sampling the best fit resulted in LUmax = 3.5* 105 ±1.5 a.u. and KD = 996±163 μM (see figure 4), standard deviations are calculated with the Jacobian.
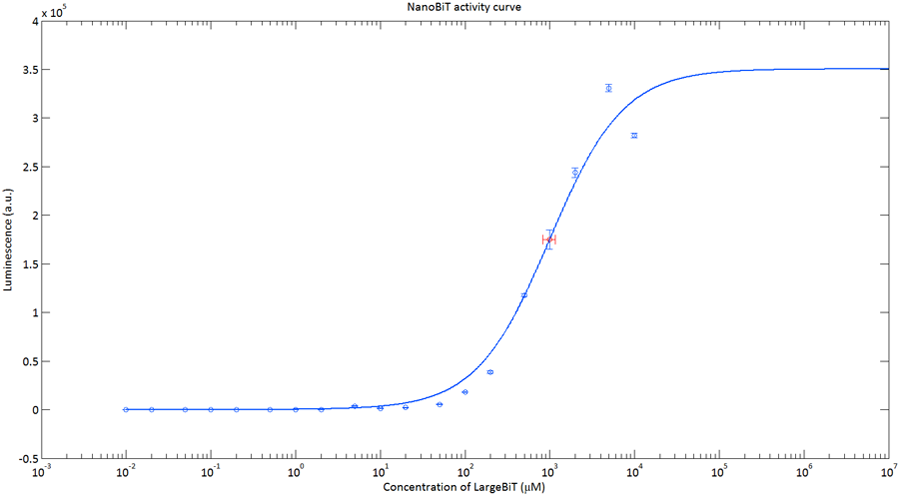
Figure 4:The Luminescence of NanoBiT as a function of the concentration of LargeBiT. The concentration of smallBiT is kept constant at 100 nM. The circles represent the measured datapoints, the errorbars represent the standard deviation of the measures (n = 2). The red circle and errorbars represent the dissociation constant and its standard deviation respectively.
Informational Contribution iGEM TU-Eindhoven 2017
The sequence has been determined and published by TU-Eindhoven 2017[http://2017.igem.org/Team:TU-Eindhoven]. The sequence does not include the ATG start codon. The end of the part contains a Stop codon, which makes this part suitable to place behind other BioBricks. The information of the sequence can allow people to acquire more insights on the functionality of the part.
Improvement by DUT_China_B 2019
A better split site was found by DUT_China_B 2019,based on their modelling.The two new split protein has higher affinity than LargeBit and SmallBit, and the luminescence intensity is 3 times higher than the previous parts.
2020 Improvement UZurich
For our 2020 iGEM project, we from the University of Zürich designed a novel approach for a biosensing system relying on plant pattern-recognition receptors (PRRs). We managed to engineer parts of the system in the lab and conduct initial experiments to characterize our system. After many discussions with experts, we decided to use the split-NanoLuc system to generate a luminescent output. Since our system was designed for our chassis S. cerevisiae we decided to use a codon optimized sequence for the output, as we wanted to facilitate expression in our chassis. For this purpose, we codon optimized this part and also Part:BBa_K1761005, which encodes for the SmallBit, the second half of the functional NanoLuc luciferase. The registry numbers for the codon optimized parts are : BBa_K3610014 for LargeBit and BBa_K3610015 for SmallBit. As we were using these parts in our project, we decided to test our improved parts and compare them with the original sequences.
Approach
Our goal was it to test whether expression of the codon optimized parts in S. cerevisiae would enhance luminescence intensity of a sample when the NanoLuc substrate furimazine is added.
We tackled this question by using rapamycin and its binding ligands, FKBP and FRB (Part:BBa_K3610022, Part:BBa_K3610023).
FKBP
he FK506 binding protein, or FKBP, is a protein belonging to the immunophilin family and has proven to be a useful tool in biological research. Functionally, the protein is a peptidyl-prolyl cis-trans isomerase (PPI).
Responsible for its prominent role in research is the ability of FKBP to bind to rapamycin, an antifungal antibiotic macrolide. What makes this interaction interesting is the fact that rapamycin binds to FKBP and the FKBP–rapamycin binding (FRB) domain of the mammalian target of rapamycin (mTOR) simultaneously, inducing a dimerization of these two components. These properties of FKBP and FRB have been exploited for conditional dimerization of proteins of interest, which offers many possibilities to artificially manipulate cellular processes, by fusing them to FKBP and FRB.
FRB
The mammalian target of rapamycin (mTOR) is a Ser/Thr protein kinase, which is involved in the control of mRNA translation initiation. The FKBP12-rapamycin binding domain (FRB) is located upstream of the catalytic domain and is essential for the kinase activity of the mTOR protein. Without the FRB domain the protein loses its functionality. As already established the interesting property of FRB is its interaction with the antifungal antibiotic macrolide rapamycin. Rapamycin binds to FRB and the FK506 binding protein (FKBP) simultaneously, inducing dimerization of the two components. This rapamycin-induced dimerization has been exploited for dimerization of proteins of interest by fusing them to FKBP and FRB.
Rapamycind binding ligands and NanoBits
The previously described properties come in handy when we need to induce protein-protein interaction. We decided to test the improved construct in a dimerization assay with rapamycin.
For this purpose, we designed the following constructs:
- Part:BBa_K3610054 LargeBit fused to FKBP12 (LBit:FKBP), Part:BBa_K3610055 optimized LargeBit fused to FKBP12 (LBit:FKBP_yeast), Part:BBa_K3610056 SmallBit fused to FRB (SBit:FRB), Part:BBa_K3610057 optimized SmallBit fused to FRB (SBit:FRB yeast).
The split protein was fused to either FKBP12 or FRB via a 15 amino acid(AA)-long linker consisting of the small AAs Glycin and Serine. Additionally, the two LargeBit constructs contain a hemagglutinin tag at between the receptor protein-coding region and the 15AA-linker, which is used for (co-)immunoprecipitation and Western Blotting. The SmallBit constructs have a FLAG tag at the same position for the same purpose.
For testing our newly created part Part:BBa_K3610014, we co-expressed LBit:FKBP and SBit:FRB in <S. cerevisiae (AP4)</i> and did the same for LBit:FKBP_yeast and SBit:FRB. When both parts are expressed, addition of rapamycin will make the ligands FKBP12 and FRB bind to it, which will initiate the interaction between the LargeBit and the SmallBit. Since SmallBit is very short and has hardly any codons that are rare in S. cerevisiae, we only tested the codon optimized LargeBit. Should codon optimization actually facilitate expression in yeast, we would expect the luminescence intensity to be higher, when rapamycin and the substrate furimazine are added.
Preparation of the constructs
In a first step we inserted the single fragments making up this part into a plasmid with a gentamycin-3-acetyltransferase gene and transformed E. coli (DH10alpha) with the plasmids for amplification. In the next step we assembled the fragments in a plasmid with a spectinomycin acetyltransferase and amplified the plasmids again in the same E. coli strain. For this step we applied the techniques of Golden Gate Cloning to get the fragments in the right order into the plasmid. The restriction enzyme we chose was BsaI.
We expressed the S. cerevisiae cells with two different plasmids, each containing one construct. One plasmid with a -TRP gene, enabling the yeast strain to produce tryptophan, the other contains a gene for the kanamycin acetyltransferase. We therefore needed a synthetic dropout medium (-TRP) supplemented by Geneticin.
Procedures
After successful transformation, we examined the cells with a plate reader of the type Synergy H1. Cells were inoculated in liquid medium for several hours at 30°C and adjusted to the same OD600. This step was necessary to prevent differences in measured values due to higher cell density in one sample.
Afterwards, the 96 well plate was prepared with 50μl samples containing LBit and samples containing LBit_yeast. We divided the samples into groups, those to which no rapamycin was added and those that received additional rapamycin. The substrate furimazine was added to each well in form of 50μl NanoGlo solution (50:1 buffer to substrate). The rapamycin was added immediately after addition of furimazine. 10μM rapamycin was added, giving a final concentration of 10 μM.
Further Details:
- Runtime 2:00:00 (HH:MM:SS)
- Interval 0:06:00, 21 Reads
- Shake
- Linear:
- Continuous
- Frequency: 493 cpm (4 mm)
- Read
- Luminescence Endpoint
- Full Plate
- Integration Time: 0:03.00 (MM:SS.ss)
- Filter Set 1
- Emission: Full light
- Optics: Top, Gain: 135
- Read Speed: Normal, Delay: 100 msec
- Extended Dynamic Range
- Read Height: 1 mm
- Linear:
Results
Assay 1
In the first measurement series, all samples were adjusted to OD600 = 0.26. The following samples were prepared:
- 6X LBit_yeast without rapamycin
- 6X LBit yeast with rapamycin
- 6X LBit registry without rapamycin
- 6X LBit registry with rapamycin
(LBit registry referres to this part)
Measurements with a luminometer at 480 nm over two hours were performed (T° Lum for each measurement between 23.9 and 24.1).
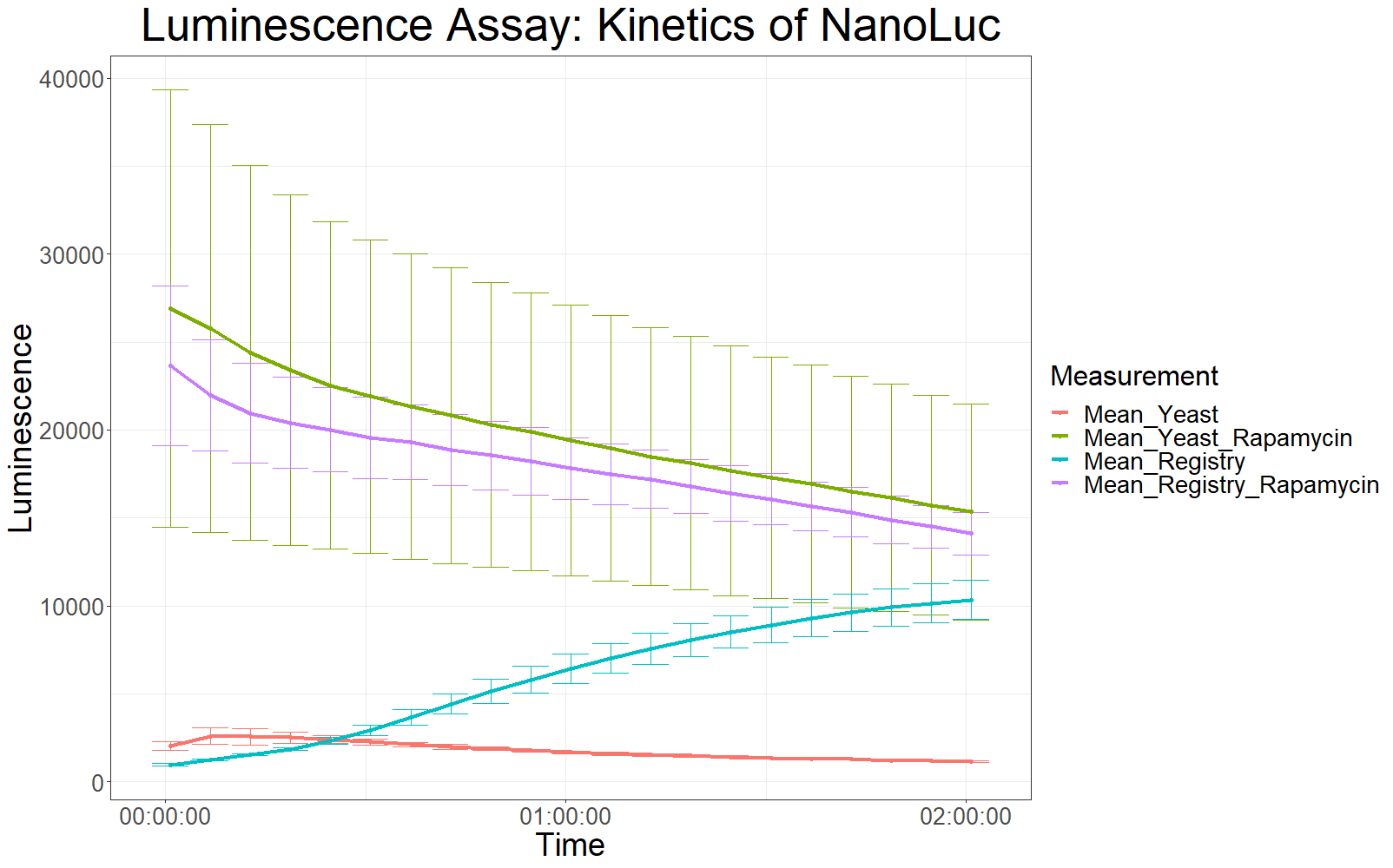
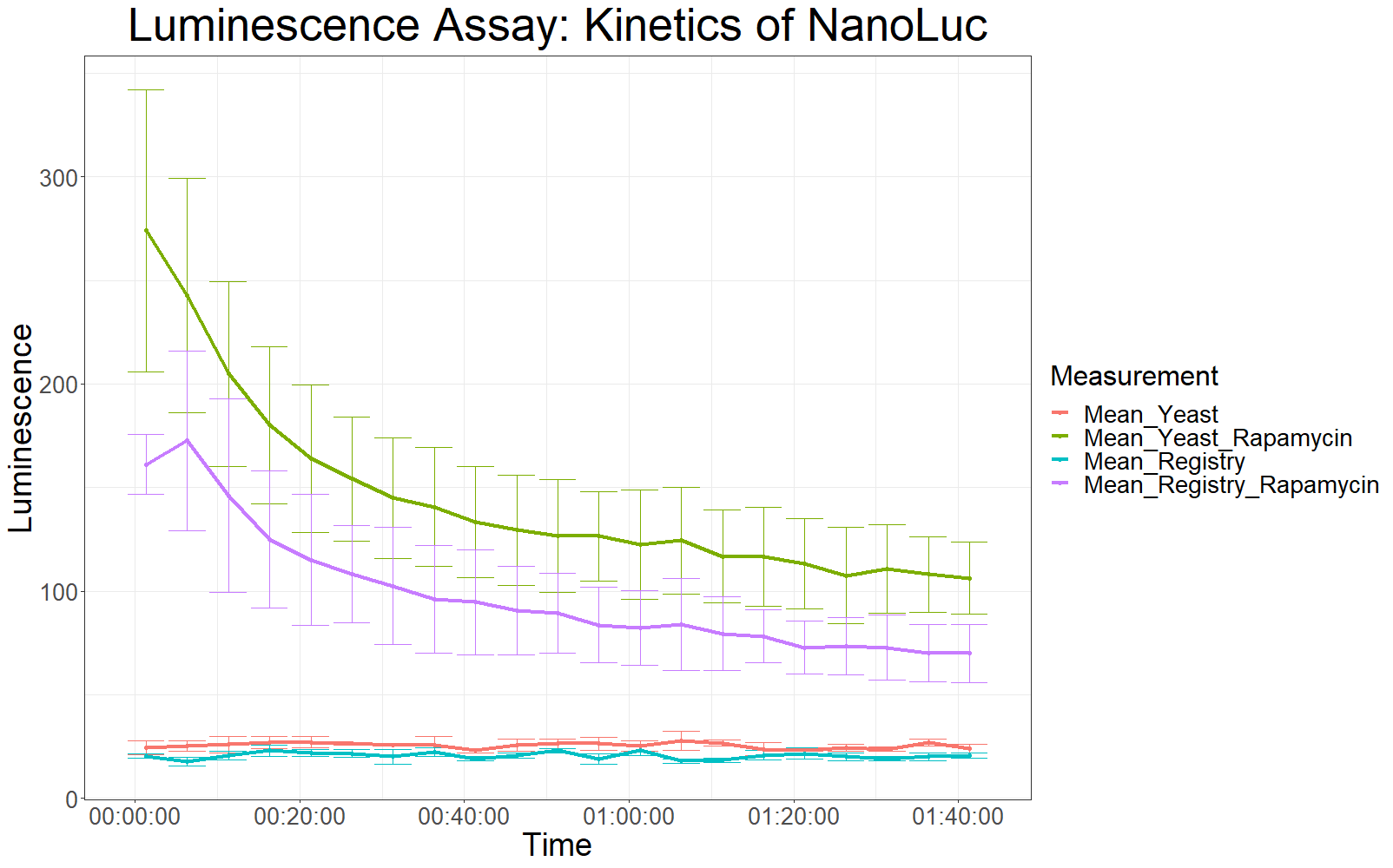
The measurement values are summarized in the graph below:
As expected, the control samples (no rapamycin added) gave hardly any detectable luminescent output. When rapamycin was added to the wells, increased luminescence intensity could be detected. This initial assay showed increased luminescence in samples transformed with LBit:FKBP_yeast (codon optimized). The variance, however, is too big to make any assumptions after this initial measurement (regarding increased luminescence when compared to the original sequence).
Assay 2
Because the first data from the first measurement was not very conclusive, the same type of measurement was repeated. With the following differences:
- OD600 = 0.5 for all samples
- longer waiting period between taking cells out of the incubator and measurement (2 hours)
- After addition of NanoGlo solution, the samples were incubated at RT for 30 minutes.
The following samples were measured:
- 3X yeast without rapamycin
- 9X yeast with rapamycin
- 3X registry without rapamycin
- 3X registry with rapamycin
Analysis
The patterns observed in the second experiment were similar to the ones observed in the first assay. Expression of the constructs was successful and rapamycin was able to drive the interaction between the LargeBit and the SmallBit part, which is made visible by the luminescent output after addition of the NanoLuc substrate furimazine.
As had already been previously observed, samples which expressed the codon optimized version of this part, showed higher average fluorescence intensity at all measurement points.
When comparing Yeast_Rapamycin and Registry_Rapamycin the overall average luminescence intensity was significantly different (W = 6669.5, p-value < 2.2e-16, Wilcoxon Ranked Signed Rank Test was conducted as the measurements were made with cells coming from the same plate).
Conclusion
We conducted initial tests to characterize the parts Part:BBa_K1761005 and Part:BBa_K3610014. When SmallBit and LargeBit are fused to the two rapamycin binding ligands FRB and FKBP12 respectively and expressed in S. cerevisiae (AP4), addition of rapamycin is able to drive the interaction between the SmallBit and the LargeBit, leading to the reconstitution of their functionality as a luciferase. Addition of rapamycin and the substrate furimazine will lead to a luminescent output.
Additionally, initial measurements with a luminometer suggested a stronger overall luminescence intensity, when the codon optimized sequence of LargeBit was used.
References
[1] Kyle Hooper, "Applications of a smaller, brighter, more versatile luciferase: NanoLuc™ Luciferase Technology", Presentation slides Fall 2012. [3]
| emission | 460 nm |