Help:Constructing Parts
- Registry Help Pages:
- TOC
- At-a-Glance
- FAQ
Constructing a new part is one of the most important steps to making a contribution to the synthetic biology community and the Registry. After adding a part to the Registry database, you'll find yourself in the lab, constructing a physical sample of that part. Maybe you're synthesizing an entirely new basic part, extracting one from another organism, or putting together a new device (composite part), we'll detail some the methods that you can use when constructing your parts in the lab.
Don't forget the Registry's part submission requirements: all parts need to be sent to us in the pSB1C3 plasmid backbone. Regardless of the standard you used during your synthetic biology project you will need to make sure that the sample you send to the Registry adheres to the BioBrick RFC[10] and is in the pSB1C3 shipping backbone.
Basic Part
DNA Synthesis
If you are constructing a part via direct synthesis, you will need make sure that there are no illegal restriction sites that would interfere with your standard's assembly. Based on the requirements for sending your part to the Registry (packaged in pSB1C3 plasmid backbone) your part will be flanked by the BioBrick prefix and suffix. Therefore the part will need to be BioBrick compatible, and cannot have the following restriction sites: EcoR1, Xba1, Spe1, Pst1.
See the DNA synthesis page for more information.
Most synthesizing companies will send your product back in their own specialized plasmid. If you are able to include the prefix and suffix, required by your assembly standard, in the synthesis of your part then you will be able to cut your part out from their plasmid and into a Registry supported plasmid backbone. Otherwise, you can use PCR to extract your synthesized part and add on the appropriate prefix and suffix.
PCR
Many interesting parts exist in nature or in your lab. You can use PCR to extract the DNA sequence to create your part and place the correct prefix and suffix on its ends. You will have to be sure that the part's sequence is compatible with the assembly standard you wish to use.
You will also find that some available part samples from the Registry are not in a plasmid backbone that belong to your specific assembly standard. You can add your preferred standard's prefix and suffix in the same manner.
Don't forget that part samples sent to the Registry, must be packaged in the pSB1C3 plasmid backbone. If you used a different assembly standard in the construction of your parts you can use PCR with BioBrick prefix and suffix primers that will allow you to transfer your part into pSB1C3.
Prefix
- 5' GTTTCTTCGAATTCGCGGCCGCTTCTAGAG[part] 3'
Suffix
- 5' [part]TACTAGTAGCGGCCGCTGCAGGAAGAAAC 3'
See the PCR Standardization page for more information.
Site-directed Mutagenesis
Site-directed mutagenesis is particularly useful if you need to remove illegal restriction sites to make your part compatible with your preferred assembly standard and BioBrick RFC[10].
Generally this involves...
- Purifying template plasmid DNA containing your part from a dam+ E. coli strain (ensures that template plasmid is methylated for digestion with DpnI).
- Designing forward and reverse primers, containing the mutation you want, that will bind to the correct region of DNA.
- Running a primer-extension reaction with a proof-reading, non-displacing polymerase such as Pfu DNA polymerase. This results in nicked circular strands of the plasmid.
- Cut up the template plasmid DNA with DpnI.
- Transform the circular nicked DNA into a highly competent cells. These cells will repair the nicks and not restrict the unmodified product DNA.
- Select colonies and test them (colony PCR, restriction digest, sequencing) to find your correct mutant.
See the Site-directed Mutagenesis page for more information.
Composite Parts
Restriction Enzyme Assembly
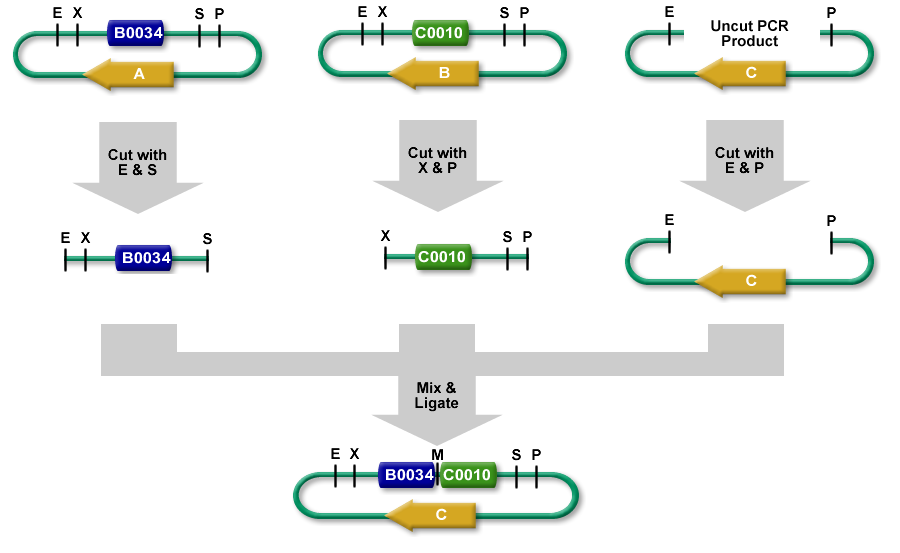
3A Assembly
3A Assembly (which stands for three antibiotic assembly) is a method for assembling two part samples and selecting for correct assemblies through antibiotics. 3A assembly uses the restriction sites on the prefix and suffix to assemble part samples. This new composite part will maintain the same prefix and suffix as its "parents" and contain a scar, where the cut and re-ligated restriction sites were stitched together.
It uses effective antibiotic selection to eliminate unwanted background colonies and eliminates the need for gel purification and colony PCR of the resulting colonies. In theory, about 97% of the colonies should be the desired assembly. Additionally, the Registry has been providing iGEM teams and Registry labs with linearized plasmid backbones to further improve this assembly method.
3A Assembly Advantages
- No PCR
- No gel purification
- Higher success rate compared to Standard Assembly.
Links
Type IIS Assembly Methods
Type IIS restriction enzymes, like BsaI, cut DNA outside of their recognition site. This allows for custom overhangs, unlike normal restriction based enzymes (EcoRI cuts at its recognition site creating the same overhang every time). Type IIS assembly methods, such as MoClo, use this to their advantage. Parts will have the same restriction enzyme but distinct custom overhangs, so multiple parts can be assembled at once (one pot assembly).
Type IIs assembly requires careful planning and preparation to ensure the multiple parts will assemble properly and in the correct order. Assembly standards like MoClo, help make this process easier by standardizing part.
Currently, the Registry is evaluating MoClo, to support it as another assembly systems.
Scarless Assembly
There are assembly methods which also allow for the construction of composite parts without scars or specific linkers, which is particularly useful for the assembly of proteins. Additionally, these methods may be able to circumvent assembly standard incompatibility between part samples: a part sample in a Silver RFC[23] plasmid backbone can be assembled with a part sample in a Berkeley RFC[21] plasmid backbone.
DNA Synthesis
Just as DNA synthesis can be used to construct a new basic part, it may also be used to assemble parts together. Using the Registry database you can pull the sequence for parts you're interested, put them together in series, and then send the new composite sequence off to be synthesized.
See the DNA synthesis page for more information and our offer with IDT.
Gibson Assembly
Gibson Assembly has not been tested by the Registry yet, but several teams have had success with this assembly method. The [http://2010.igem.org/Team:Cambridge/Gibson/Introduction Cambridge 2010 iGEM Team] developed a set of protocols and tools that may be useful.
The Registry will be evaluating Gibson Assembly, and will have resources available for this assembly method available soon.
Discontinued Assembly Methods
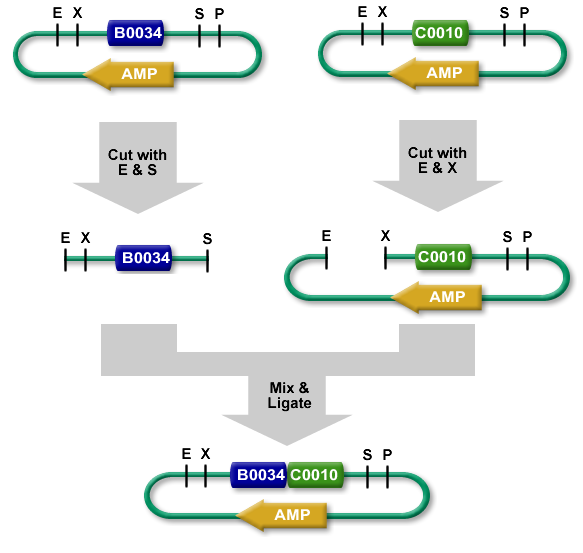
Standard BioBrick Assembly
For Standard BioBrick Assembly, a part sample is cut out from its plasmid backbone and inserted into the prefix of a plasmid backbone of another part. Two restriction digests are done, one for the part sample that will be moved and one for the plasmid backbone that will receive it. The digests are then run on a gel and using gel purification the required fragments are isolated (the part sample and the cut plasmid backbone). The purified insert and cut plasmid backbone are ligated and the resulting composite part can be transformed into E.coli cells.
Due to its use of gel purification and lower success rate, the Registry no longer recommends the Standard BioBrick Assembly method.
Links
I've constructed a part, now what?
Now it's time to use and characterize it! The documentation that is provided along with your part will allow users in the Registry community to better understand the part before they use it.
Make & Give...