Part:BBa_K525222
S-layer cspB from Corynebacterium halotolerans
S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self assembly-domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to eachother (determined by the S-layer geometry) it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr et al., 2007]).
Usage and Biology
S-layer proteins can be used as scaffold for nanobiotechnological applications and devices by e.g. fusing the S-layer's self-assembly domain to other functional protein domains. It is possible to coat surfaces and liposomes with S-layers. A big advantage of S-layers: after expressing in E. coli and purification, the nanobiotechnological system is cell-free. This enhances the biological security of a device.
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression (E. coli) | Compatibility | E. coli KRX |
| Induction of expression | L-rhamnose for induction of T7 polymerase | |
| Specific growth rate (un-/induced) | 0.245 h-1 / 0.109 h-1 | |
| Doubling time (un-/induced) | 2.83 h / 6.33 h | |
| Characterization | ||
| Number of amino acids | 460 | |
| Molecular weight | 50.7 kDa | |
| Theoretical pI | 4.25 | |
| Localization | cytoplasm | |
| partially in culture supernatant | ||
| partially periplasm | ||
| not cell membrane |
|} </center>
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1103
Illegal XhoI site found at 559 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 226
Illegal NgoMIV site found at 1315
Illegal AgeI site found at 217
Illegal AgeI site found at 458
Illegal AgeI site found at 505 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 907
Illegal BsaI.rc site found at 214
Illegal BsaI.rc site found at 592
Illegal BsaI.rc site found at 994
Expression in E. coli
For characterizations, the cspB gene was fused to a monomeric RFP (BBa_E1010) using Gibson assembly.
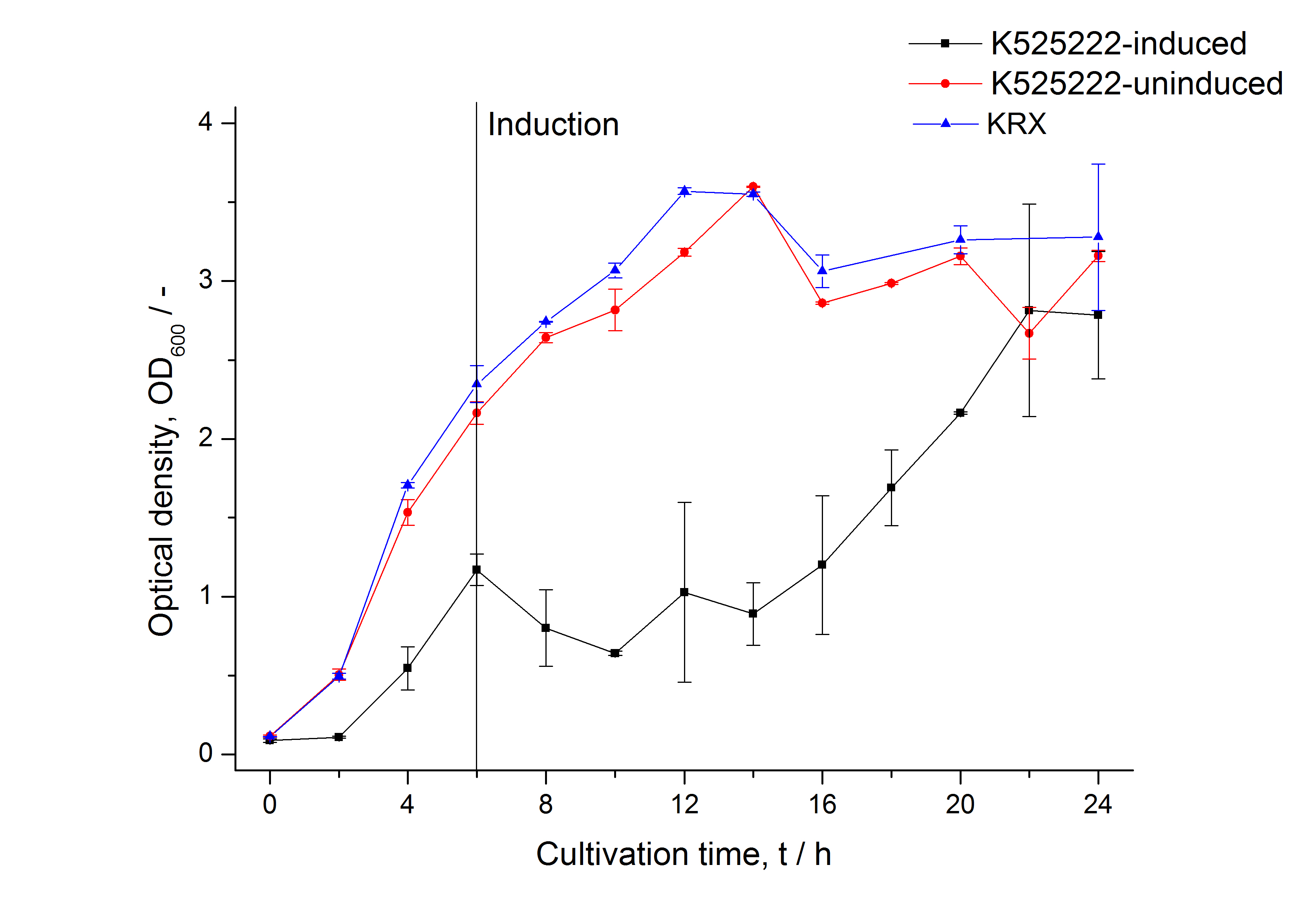
The CspB|mRFP fusion protein was overexpressed in E. coli KRX after induction of a T7 polymerase gene in the KRX's genome by supplementation of 0.1 % L-rhamnose using the autinduction protocol developed by Promega.
Identification and localisation
After a cultivation time of 18 h the mRFP|CspB fusion protein was localized in E. coli KRX. Therefore a part of the produced biomass was mechanically disrupted and the resulting lysate was washed with ddH2O. Then the lysate was treated with ionic, nonionic and zwitterionic detergents to release the mRFP|CspB out of the membranes, if it integrates. From the other part of the cells the periplasm was detached by using an osmotic shock.
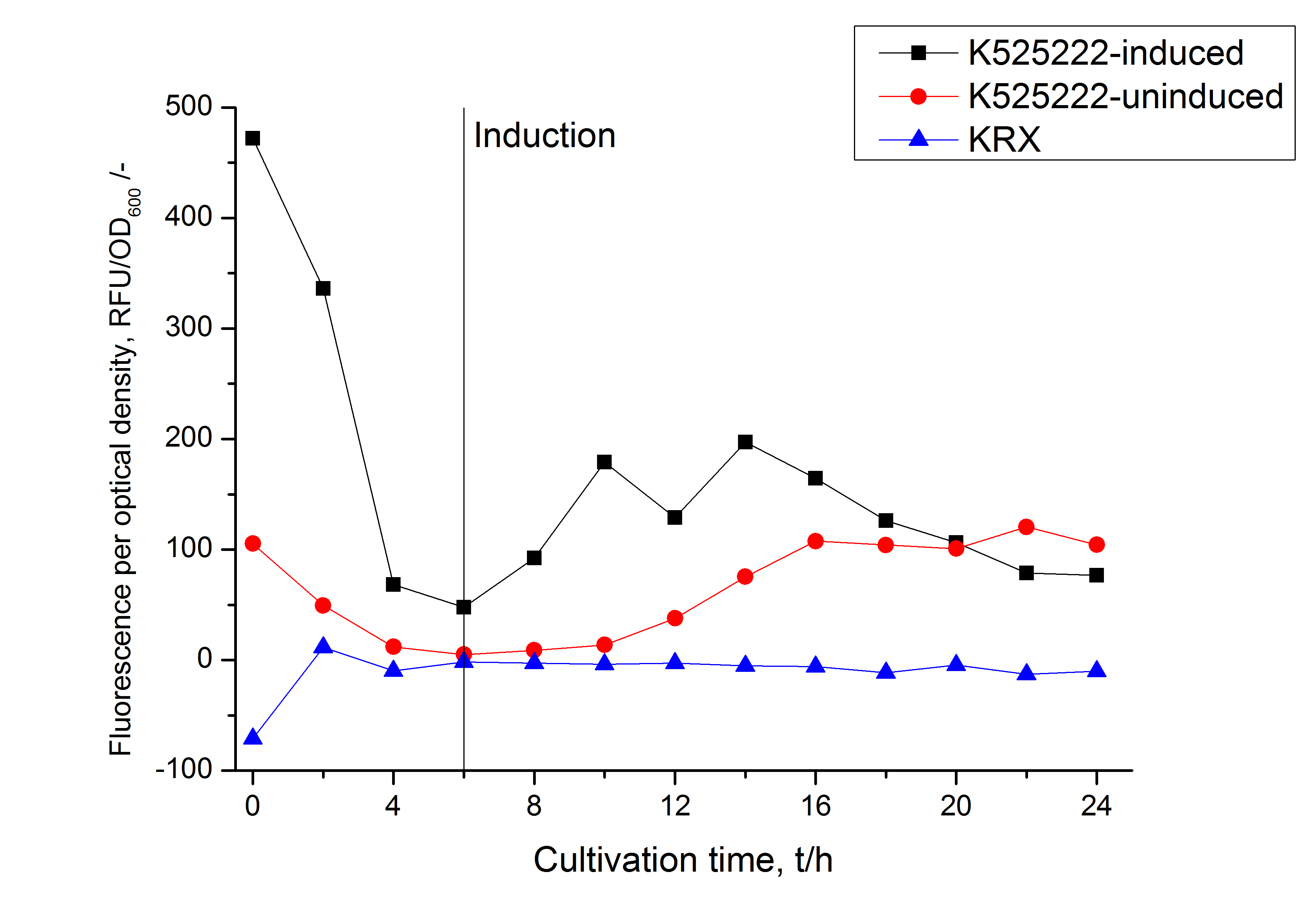
The fluorescence in all cultivation fractions plus the fluorescence in the lysis und wash fraction shows that the fusion protein is water soluble and does not sediment during centrifugation. Together with the absence of flourescence in the detergent fractions this verifies that the fusion protein is not integrated into the cell membrane (fig. 3) and is not forming inclusion bodies.
In comparison with the mRFP fusion protein of BBa_K525224, which has a TAT-sequence, a minor relative fluorescence in all cultivation and detergent fractions was detected (fig. 3). Together with the decreasing RFU after 14 h of cultivation (fig. 2) this result indicates a postive effect of the TAT-sequence on the protein stability. This could be due to a digestion of BBa_K525222 by proteases in the cytoplasm.
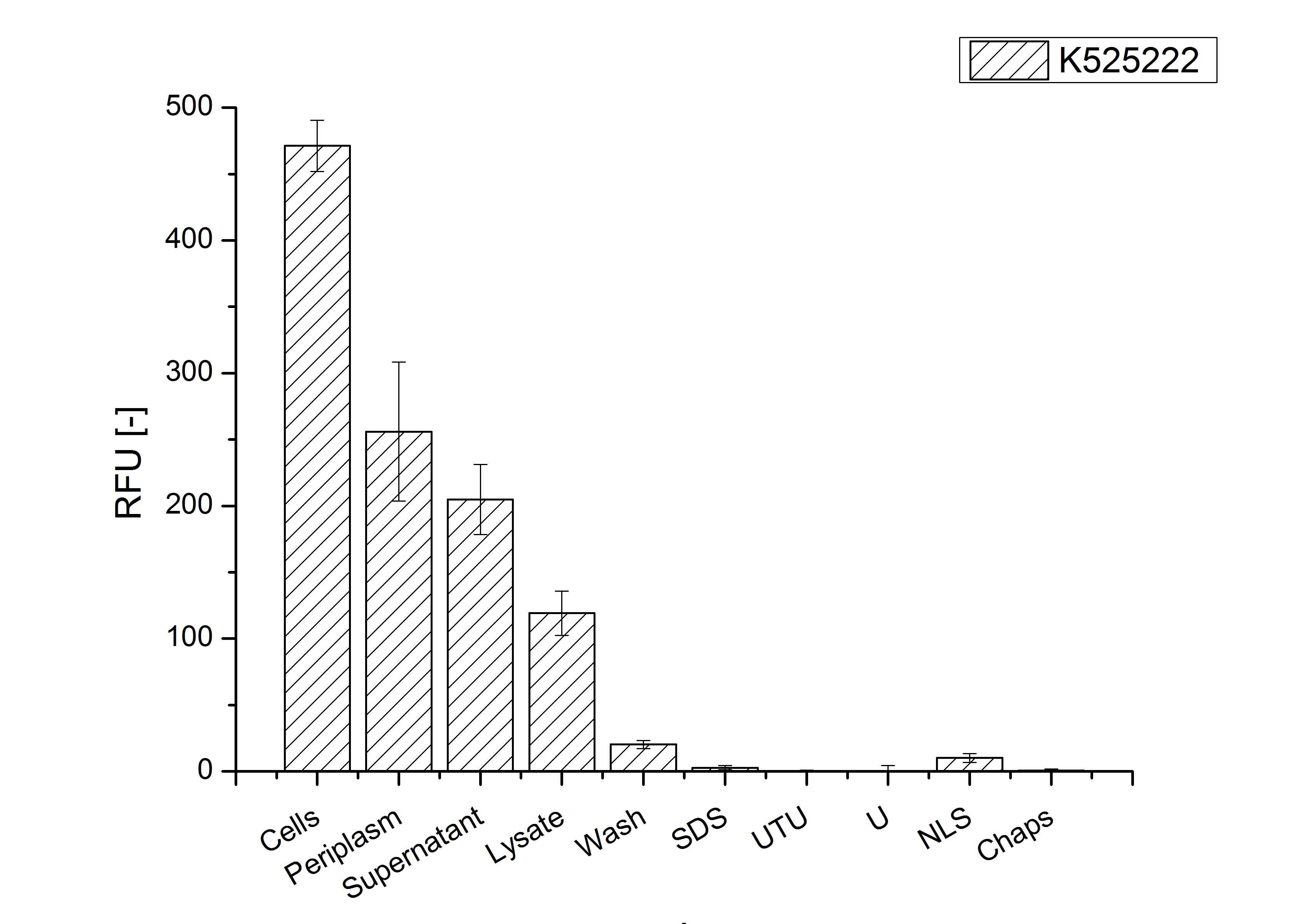
MALDI-TOF analysis was used to identify the location of the fusion protein in different fractions. Fractions of medium supernatant after cultivation, periplasmatic isolation, cell lysis and following denaturation in 6 M urea were loaded onto a SDS_PAGE. After denaturation with 6 M urea the remaining pellet (after centrifugation 15,000 g for 30 min) was washed with 2 % (v/v) Triton X-100, 2 % SDS (w/v). This fraction was also loaded onto the SDS-PAGE and fragments of the gel were measured with MALDI-TOF.
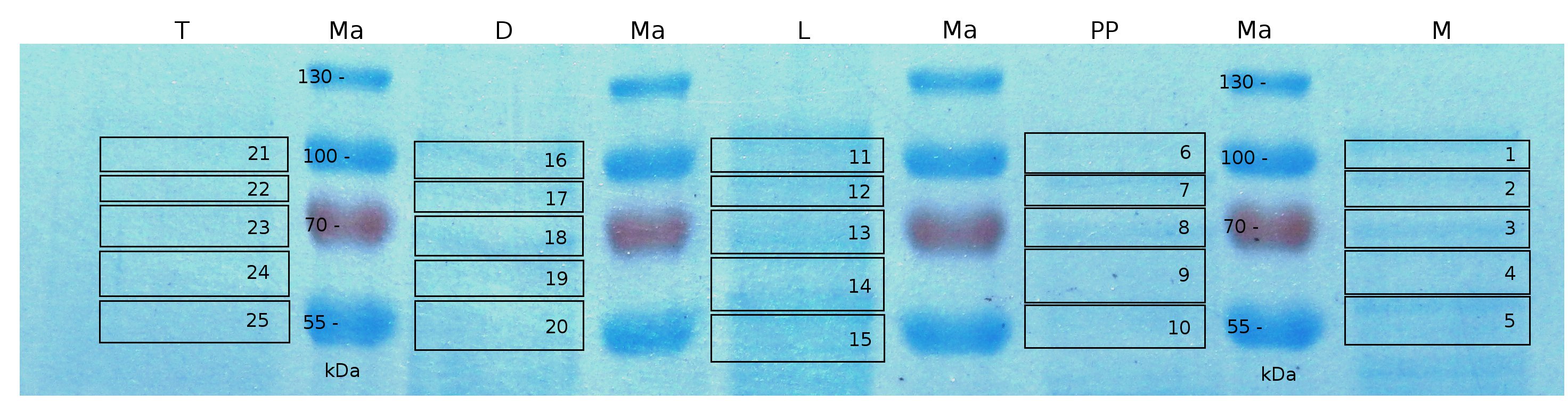
The following table shows the sequence coverage (in %) of our measurable gel samples with the amino acid sequence of fusion protein CspB|mRFP (BBa_E1010).
| number of gel sample | sequence coverage (%) |
|---|---|
| 1 | 0.0 |
| 2 | 0.0 |
| 3 | 0.0 |
| 4 | 0.0 |
| 5 | 0.0 |
| 6 | 0.0 |
| 7 | 0.0 |
| 8 | 0.0 |
| 9 | 0.0 |
| 10 | 0.0 |
| 11 | 0.0 |
| 12 | 14.6 |
| 13 | 9.3 |
| 14 | 6.3 |
| 15 | 0.0 |
| 16 | 0.0 |
| 17 | 0.0 |
| 18 | 0.0 |
| 19 | 0.0 |
| 20 | 1.0 |
| 21 | 0.0 |
| 22 | 0.0 |
| 23 | 0.0 |
| 24 | 0.0 |
| 25 | 0.0 |
Fig. 5 shows these data. The gel samples were arranged after estimated molecular mass cut out from the gel.
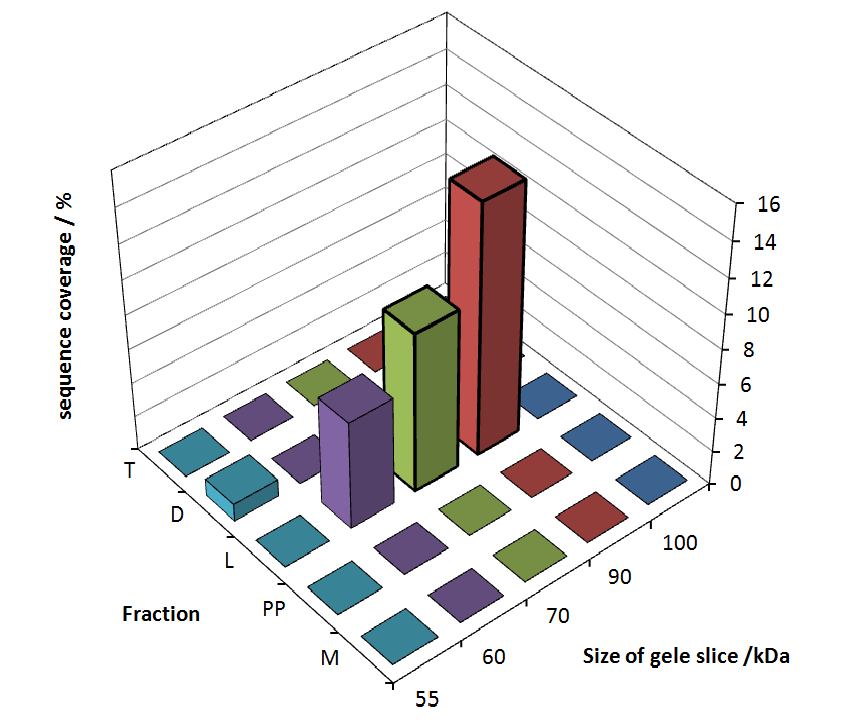
As expected, no sequence coverage was found in the periplasmatic fraction, due to absent TAT-sequence located at the amino-terminus. Low sequence coverage was found in the fraction of supernatant of the media, indicating that the protein can not be transported to the periplasm and thus secretion into the medium does not take place. The denaturation fraction and the Triton X-100 fraction show no or very few sequence coverage, however the lysis fraction shows significant higher sequence coverage. Both results indicate, that the fusion protein is solely present in the cytoplasm and thus only identified in the lysis fraction. Fig. 5 and fig. 6 show that the protein can be found mainly in the lysis fraction, but in smaller amounts in the periplasmatic and the media fraction as well, which can be explained due to the abscence of the lipid anchor. The anchor normally binds to the cell membrane, so no protein is found in other fractions than the lysis fraction.
To obtain more specific informations about the location of the S-layer fusion protein, after comparison with same treated fraction of E. coli KRX all gel bands in a defined size area were cut out of the gel and analysed with MALDI-TOF. Results are shown in fig. 6.
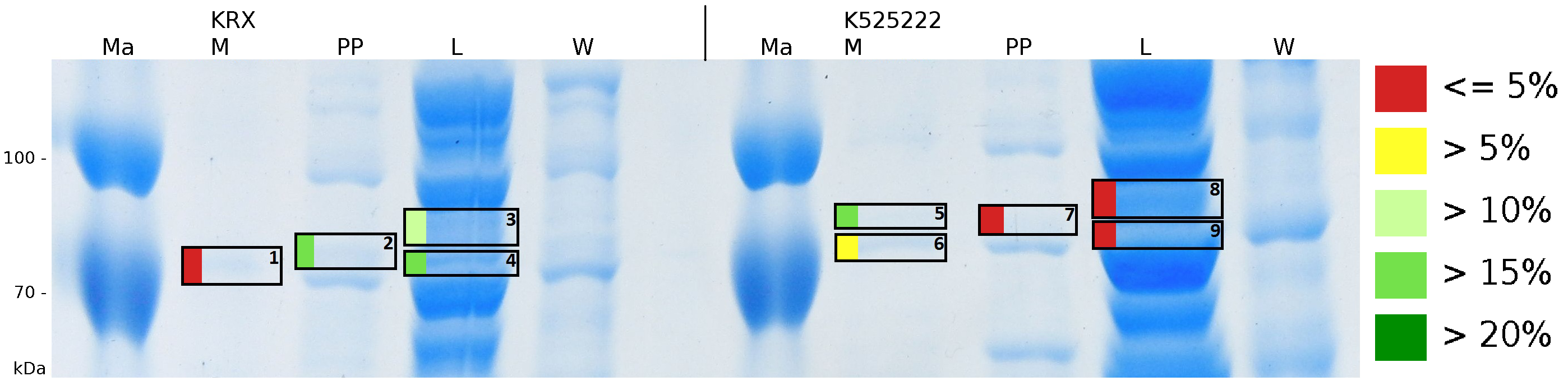
MALDI-TOF measurement shows sequence coverage in the supernatant fraction of the cultivation, the periplasmatic fraction and the lysis fraction, indicating that the fusion protein of BBa_K525222 and BBa_E1010 without TAT-sequence and lipid anchor is not only found in the cytoplasm. The periplasmatic isolation does destroy a few complete cells, which could leed to the sequence coverage found in the periplasmatic fraction. The gel lanes in the periplasmatic fraction are clearly less intense, the protein concentration is very low compared to the lysis fraction. Sequence coverage in the media fraction shows, that the protein is probably secreted into the media. Cell lysis could leed to this effect. The sequence coverage in a MALDI-TOF analysis does not automatically correlate with protein concentrations. Small amounts of proteins can give the same results when cutting out a clean protein band of a single protein. When looking at the intensity of the analysed gel pieces it can be summarized, that there is some CspB found in the periplasm and the culture supernatant but most of the protein stays in the cytoplasm which was the expected result. No fluorescence and sequence coverage was found in the cell membrane fractions, giving another proof that the lipid anchor from Corynebacterium binds to the cell membrane of E. coli.
Methods
Expression of S-layer genes in E. coli
- Chassis: Promega's [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ E. coli KRX]
- Medium: LB medium supplemented with 20 mg L-1 chloramphenicol
- For autoinduction: Cultivations in LB-medium were supplemented with 0.1 % L-rhamnose as inducer and 0.05 % glucose
Measuring of mRFP
- Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination
- Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark
- To measure the samples thaw at room temperature and fill 200 µL of each sample in one well of a black, flat bottom 96 well microtiter plate (perform at least a repeat determination)
- Measure the fluorescence in a platereader (we used a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]) with following settings:
- 20 sec orbital shaking (1 mm amplitude with a frequency of 87.6 rpm)
- Measurement mode: Top
- Excitation: 584 nm
- Emission: 620 nm
- Number of reads: 25
- Manual gain: 100
- Integration time: 20 µs
Tryptic digest of gel lanes for analysis with MALDI-TOF
Note:
- Make sure to work under a fume hood.
- Do not work with protective gloves to prevent contamination of your sample with platicizers.
Reaction tubes have to be cleaned with 60% (v/v) CH3CN, 0.1% (v/v) TFA. Afterwards the solution has to be removed completely followed by evaporation of the tubes under a fume hood. Alternatively microtiter plates from Greiner® (REF 650161) can be used without washing.
- Cut out the protein lanes of a Coomassie-stained SDS-PAGE using a clean scalpel. Gel parts are transferred to the washed reaction tubes/microtiter plate. If necessary cut the parts to smaller slices.
- Gel slices should be washed two times. Therefore add 200 µL 30% (v/v) acetonitrile in 0.1 M ammonium hydrogen carbonate each time and shake lightly for 10 minutes. Remove supernatant and discard to special waste.
- Dry gel slices at least 30 minutes in a Speedvac.
- Rehydrate gel slices in 15 µL Trypsin-solution followed by short centrifugation.
- Gel slices have to be incubated 30 minutes at room temperature, followed by incubation at 37 °C over night.
- Dry gel slices at least 30 minutes in a Speedvac.
- According to the size of the gel slice, add 5 – 20 µL 50% (v/v) ACN / 0,1% (v/v) TFA.
- Samples can be used for MALDI measurement or stored at -20 °C.
Trypsin-solution: 1 µL Trypsin + 14 µL 10 mM NH4HCO3
- Therefore solubilize lyophilized Trypsin in 200 µL of provided buffer and incubate for 15 minutes at 30 °C for activation. For further use it can be stored at -20 °C.
Preparation and Spotting for analysis of peptides on Bruker AnchorChips
- Spot 0,5 – 1 µL sample aliquot
- Add 1 µL HCCA matrix solution to the spotted sample aliquots. Pipet up and down approximately five times to obtain a sufficient mixing. Be careful not to contact the AnchorChip.
Note: Most of the sample solvent needs to be gone in order to achieve a sufficiently low water content. When the matrix solution is added to the previously spotted sample aliquot at a too high water content in the mixture, it will result in undesired crystallization of the matrix outside the anchor spot area.
- Dry the prepared spots at room temperature
- Spot external calibrants on the adjacent calibrant spot positions. Use the calibrant stock solution (Bruker’s “Peptide Calibration Standard II”, Part number #222570), add 125 µL of 0,1% TFA (v/v) in 30% ACN to the vial. Vortex and sonicate the vial.
- Mix the calibrant stock solution in a 1:200 ratio with HCCA matrix and deposit 1 µL of the mixture onto the calibrant spots.
References
Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM (2007) S-layers as a tool kit for nanobiotechnological applications, [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full FEMS Microbiol Lett 267(2):131-144].
//proteindomain/internal
| n/a | S-layer cspB from Corynebacterium halotolerans |