Part:BBa_K4439013
mSA-GFP-CBD-10xHis
Abstract
For the scope of our project, to have a proper control for our experiments, we engineered a GFP fusion protein following the same construction as for the silk fusion protein (Part:BBa K4439007). On one hand, by fusing the sfGFP protein sequence to a Cellulose Binding Domain (CBD) on the C-terminus, the recombinant protein is able to bind the cellulose aerogel. On the other hand, by adding a streptavidin monomer (mSA) to the N-terminus, we can link this GFP fusion protein with any other protein with interesting properties via the well known biotin-streptavidin interaction. Finally, to purify the resulting protein from BL21(DE3) E. Coli competent cells, the expression system we chose, we added a 10xHis tag to the C-terminus, after the CBD. Indeed, we used this essential part to have a visual control for each step of the protein production, as well as for the coating of the aerogel, to characterize the CBD and to compare the hydrophobicity added by the silk for the proof of concept of our project.
Sequence and features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Protein Characterization
Usage and Biology
mSA
mSA, or monomeric streptavidin, is an engineered streptavidin/rhizavidin hybrid that binds to biotin with high affinity as a monomer, it was constructed and characterized by Lim et al. in 2013 (ref). It can be fused as a genetic tag to heterologous proteins to enable biotin binding. The biotin affinity of mSA is the highest among nontetrameric streptavidin, the monomer also has significantly higher stability and solubility than all other previously engineered monomers to ensure the folding and functionality of the molecule during its application. It is therefore a useful tool for biotechnology applications making use of the streptavidin-biotin interaction.
Cellulose-Binding Domain
- The Imperial 2014 iGEM team (Part:BBa K1321014) documented a particular cellulose-binding domain (CBDCipA). The Puiching Macau 2020 iGEM team (Part:BBa K3503004) used their records to fuse the CBDCipA to the SR protein, another protein that we also used in our project. In addition, the Linkoping Sweden 2019 iGEM team (Part:BBa K3182001) pursued the characterisation of CBDCipA. As this particular cellulose binding domain originates from a thermophilic bacteria (which further increases the domain's applications) and was well documented on the iGEM Parts Registry, we decided to build upon the work of those three iGEM teams.
- For the scope of our project, we reused the useful composite part CBD-SR from PuiChing Macau 2020 as it was and implemented their CBD sequence in our designs.
Modeling
Protein Modeling
We also designed a control to prove the effective attachment of our recombinant proteins.
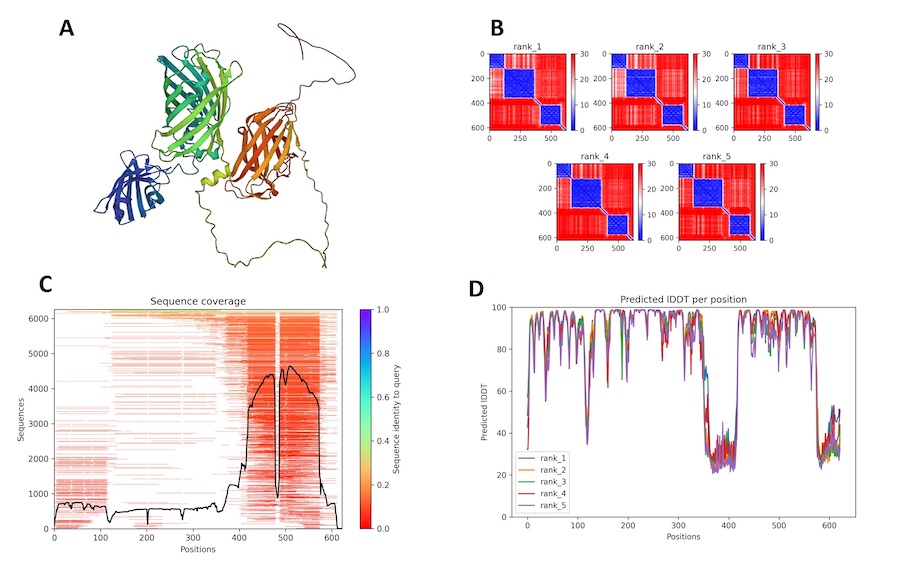
Figure 1 | AlphaFold2 prediction for 03a. (A) First rank 3D model prediction of 03a protein; the iteration who got the highest score in the modeling. (B) Different correlation graphs between the query sequence of 03a and the predicted one, for each proposed 5 models. (C) Figure of sequence coverage of 03a and indices on alignment with other sequences in the mSA. (D) IDDT graph for 03a per residue to get an idea of the confidence in the model in predicting.
- Analysis
In (fig.1, A), we could identify the three main parts of the mSA-GFP-CBD protein. Compared to the previous (fig.1, A) we could see that the GFP domain, in green, is more easily characterized and would yield a closer linkage to the mSA chain, in blue. So we will have both of the proteins closely associated. However for the CBD region, we identified a longer linkage which would mean different configuration of tha attachment possible. From (fig.1, B, D) it was really interesting to see that the 5 predictions yielded similar correlation measures and similar IDDT scores, which enabled us to confirm that the model is robust in the determination of three chain structures. The linkage would give in reality more freedom of placement of those chains.
Modeling the amount of proteins to coat a cellulose aerogel
We had different parameters that needed to be considered and we listed the most of them in (Table. 1)

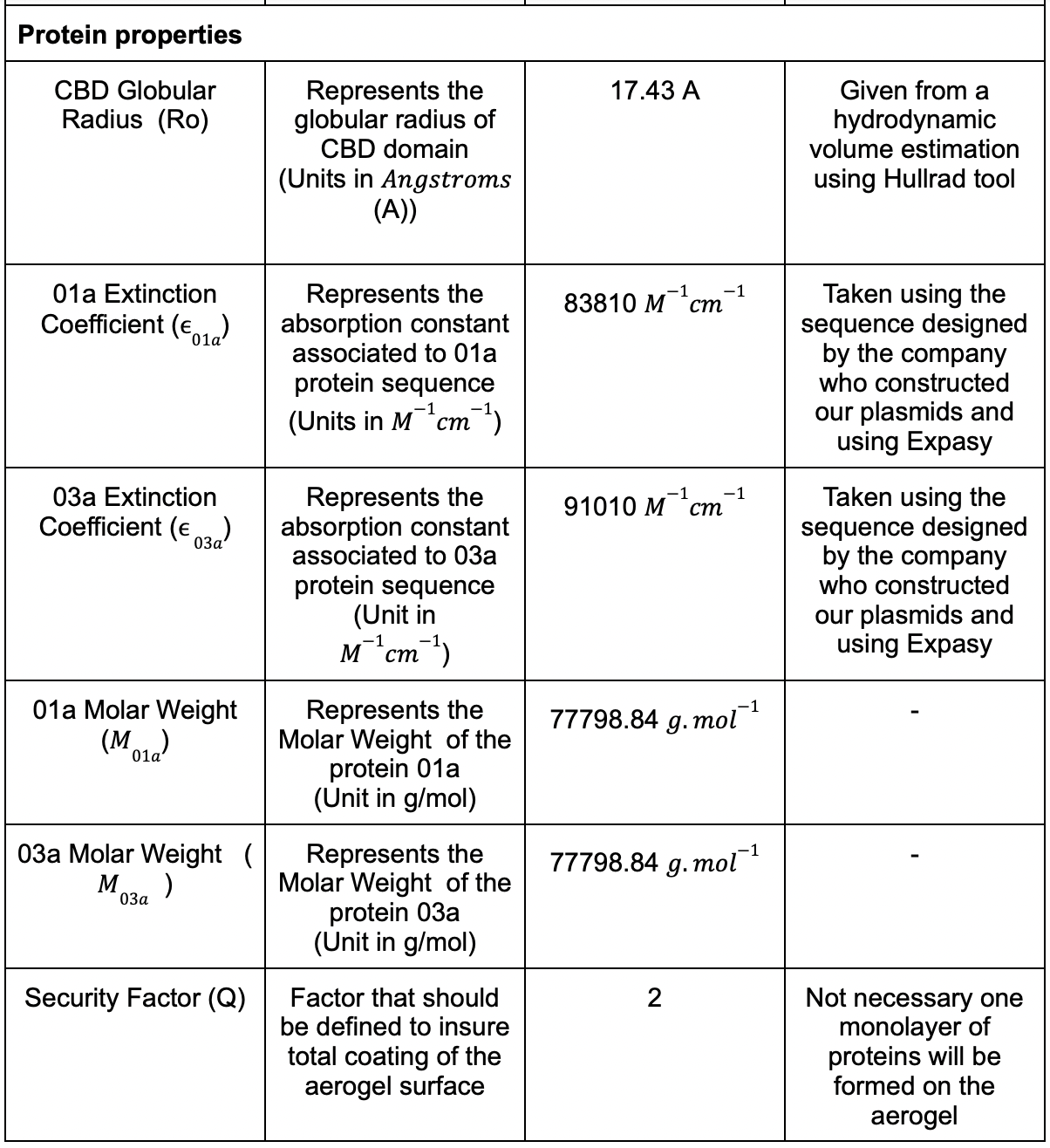
Table 1: Table for the parametric model .
In the proposed model we would like to coat geometrically our aerogel surface with different CBD globular entities. Since they are the most probable point of anchoring they will represent an entire protein in the counting of the amount of proteins. We supposed that the proteins would be placed in an adjacent manner.
First step was to consider the aerogel as a perfect cylinder that will be coated by the proteins. The surface of the cylinder is simplified to two circles and an elongated rectangle (fig2). We proceeded to a geometrical filling of the surface.

Figure 2 | Aerogel simplified representation as a perfect cylinder.
The attaching CBD region was taken from the above model 4JO5 to study its hydrodynamic characteristics using the Hullrad algorithm. This program was used to get specific useful information on the protein 4JO5:

Table 2 | Hullrad computation for CBD, AJO5
Using the values from (Table. 2), we populated the different surfaces of the aerogel with circular proteins using some simple geometry equations. The two values of AVSR and MD were studied in the example. Considering the protein radius and aerogel diameter, we came up with an iterative algorithm in three steps: Step 1: Populate the outer peripheral of the bigger cylinder:
We used the following angular equation to compute the angle of placement of the first small protein circle in the aerogel bigger circle:
alpha = 1 /π |arcsin(r(proteins)/(r(aerogel)- r(proteins)))*180| (1)
where r(proteins) and r(aerogel) represent respectively the radius of the proteins and the aerogel.
Step2: Compute the number of cylinders that can populate the peripheral of our the bigger circle:
nb(circles) =[360/(2*alpha)] (2)
where [x] is the greatest integer function s.
Step 3: Define a secondary circle that has the next small circles:
We iterated the new radius until we could not fit any small circles in the area (fig.12):
r(new)=r(previous)-( 2 *r(small)) (3)
where r(previous) is the large radius of the previous iteration.
Ending condition :
r(previous)< r(small)
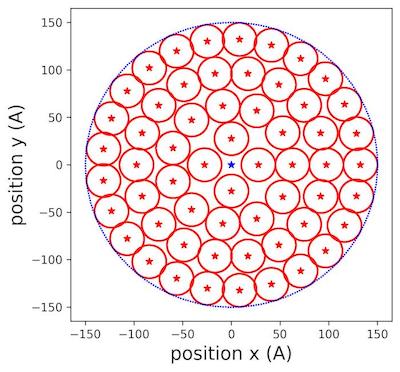
Figure 3: Populating circles with tiny cylinders zoom at 150 Å aerogel surface .
In (fig.3), we saw clearly how the different circles were placed inside the allocated surface of aerogel. We extended the reasoning to filling the rectangular lateral surface.
For the lateral specific surface, the operation is described as following:
Step 1: Divide the width of the rectangle by the number of small protein circles that fits in the line :
nb(circles for width) = [W2 * r(proteins)] (4)
where W is the width of the rectangle, r(proteins) represents the radius of the proteins and [x] is the greatest integer function as described above.
Step 2: Multiply the above number by the number of lines that can fit in the height of the rectangle:
nb(circles for rectangle) = [h2* r(small)] * nb(circles for length) (5)
where h represents the height of the rectangle, [x] is the greatest integer function as described above.
By taking the parameters of the aerogel from (Table.1) and the parameters of the CBD from (Table.2) we got the following output in number of molecules and in number of molar:

Table 3: Results of the coating computations.
- The values found in (Table.3) were useful to define the protein solution volume that we soaked the hydrogels or the aerogels with.
Results
Bacterial Transformation
We performed this bacterial transformation following the E.coli Competent Cells Quick Protocol FB035 from Promega. We transformed BL21(DE3) cells with our pET28a plasmid backbone containing mSA-GFP-CBD-10xHis coding sequence.

Figure 3 | Bacterial transformation with the 03a construct in BL21(DE3) competent cells. Remarks: unfortunately we got rid of the Rosetta plate and the control plates thinking that we took a picture of them, but it was not the case, so we do not have a picture of the empty plate and transformation controls.
- Analysis
After the overnight incubation, we observed the presence of colonies only in the BL21(DE3) transformed cells plate, but not in the Rosetta transformed cells plate. The negative control plate did not show any colony, and the positive control plate was full of colonies, as expected. The bacterial transformation of 03a worked for BL21(DE3) strain, so we can pursue the process by doing colony picking in a sterile environment (with a flame).
Protein Purification
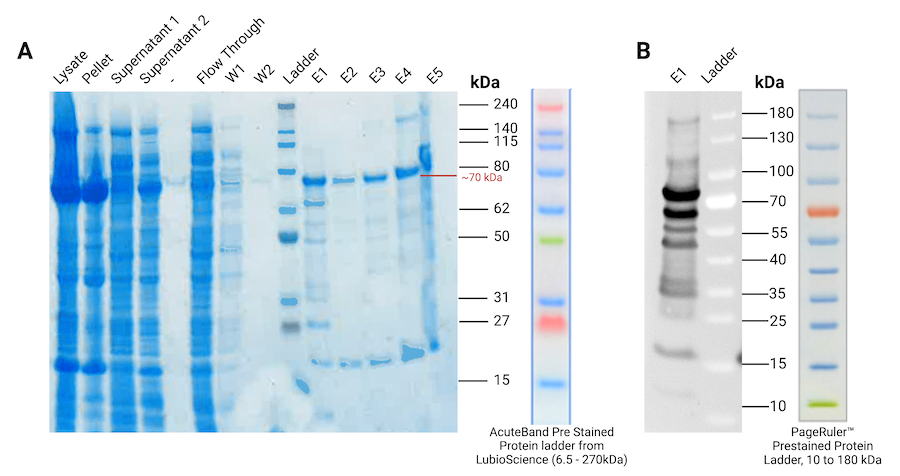
Figure 4 | Protein purification of the GFP fusion protein (03a). (A) SDS-PAGE gel stained with Coomassie Blue Protein Stain of all the fractions of GFP fusion protein purification (B) Western Blot of the elution 1 fraction (E1 on SDS-PAGE) visualised with anti-His antibody. W1 = Wash 1 with 20 mM imidazole; W2 = Wash 2 with 50 mM imidazole; E1 = Elution 1 with 250 mM imidazole; E2 = Elution 2 with 500 mM imidazole; E3 = Elution 3 with 1M imidazole; E4 = 2.5 M imidazole; E5 = 5 M imidazole.
- Analysis
On both the SDS-PAGE and the Western Blot for the protein purification of the GFP fusion protein (03a) (fig 4), we observed a band around 70kDa which matched the expected size of our protein (71.4 kDa). Purified GFP fusion protein (03a) was present in all eluted fractions. To concentrate the proteins, we used a 30 kDa filter. However, the filter was made of cellulose, so we observed the filter turning green. This showed the efficiency of the CBD domain of our construct to bind cellulose. We finally obtained a concentration of 0.35 mg/mL, which corresponds to 4.5 mg of GFP fusion proteins. Since high imidazole concentration might denature the proteins, we had to either remove it or make imidazole inert. It was not possible to dialyse the proteins to remove imidazole, since the dialysis membrane was made of cellulose, and our fusion proteins would all bind to the membrane via their cellulose binding domain (CBD). We therefore decided to flash freeze the proteins and store them in the elution buffers so the imidazole will no longer affect them when frozen.
Cloning by PCR and KLD
The elution of the proteins of interest in such high amounts of imidazole certainly comes from the double His-tag that composes our proteins. To remove it from the received plasmids, we performed some cloning experiments to obtain better yields in the future purifications. In the case of the 03a plasmid (containing GFP) and the 01b plasmid (containing SR), we performed KLD cloning. We amplified the plasmids by PCR without the undesired sequence, and we re-ligated them back by doing a KLD. The KLD reaction allows efficient phosphorylation, intramolecular ligation and template removal in a single 5-minute reaction step at room temperature.

Figure 5 | Cloning experiment results for removal of the added site for 01b (SR) and 03a (GFP). (A) Agarose gel electrophoresis of PCR products of the 01b and the 03a constructs plasmid amplified without the GeneScript additional tags for KLD cloning. (B) Agarose gel electrophoresis for restriction analysis of the 01b and the 03a constructs plasmid after the ligation by KLD. (C) Plasmid map from the sequencing result of the obtained new SR plasmid. (D) Plasmid map from the sequencing result of the obtained new GFP plasmid.
- Analysis
On the agarose gel we ran with the PCR products (fig 5.A), we observed bands around 7000 bp which are the expected sizes and means that we successfully removed the added site. After the KLD, we ran a second agarose gel with fragments obtained by cutting the new plasmids with different restriction enzymes (fig 5.B). We obtained similar patterns as the ones expected. By purifying and sequencing the plasmids(fig 5.C & D), we obtained the exact same sequence as designed. This confirmed that we removed the added site and re-ligated the DNA to obtain the good plasmids.
With several minipreps we obtained big quantities of these new plasmids. We therefore transformed new BL21(DE3) E.Coli competent cells to start the protein production. For the GFP fusion protein, the pellet after the first centrifugation step was not green, which means that there was no expression of our protein of interest. In a similar way, we couldn’t see a significant protein expression after the IPTG induction for the SR fusion protein. Even if the purification was performed, nothing would be visible in the elution lanes of the SDS-PAGE which suggests that we didn’t successfully purify our new SR fusion protein.
The cloning with PCR amplification and KLD for ligation was a success, but the bacteria were not able to produce our fusion proteins. Our hypothesis is that the number of bp between the RBS and the first Methionine was not optimal for the bacteria. However, by lack of time, we decided not to troubleshoot the reasons for this failure, and we concentrated on the protein production to use them in our further experiments.
CBD Characterisation
After the production of our proteins, we decided to test the binding of our cellulose binding domain to cellulose. We therefore added drops of our mSA-GFP-CBD protein or of a GFP that doesn’t contain CBD on a nitrocellulose membrane (fig 8A). We then performed several washes using PBS to wash out the proteins that were not bound to the membrane.

Figure 6 | Comparisons of fluorescence intensity of control GFP and our GFP fusion protein (mSA-GFP-CBD) on nitrocellulose. (A) Two samples under UV after the first PBS’ wash. (B) Two samples under UV after the third PBS’ wash. (C) BoxPlot of the difference of fluorescence intensity before and after the PBS wash for the two samples. Top of the boxes = mean of fluorescence difference per group; Black dots = individual measurements; Red vertical line = standard error of the mean. Data are mean ± s.d., n = 5 measurements per group.
- Analysis
Our mSA-GFP-CBD protein was more fluorescent than the GFP control one, even if the concentrations were the same. After the third PBS wash (fig 6B), we observed a diminution in the intensity of the fluorescence of the control GFP, while no change in intensity for the mSA-GFP-CBD. Using ImageJ, we quantified the difference in fluorescence intensity between before (fig 6A) and after (fig 6B) the PBS wash. In the case of our mSA-GFP-CBD protein, the mean for the difference in fluorescence intensity is smaller than for the control GFP (fig 6C). With this quantification, we concluded that our protein mSA-GFP-CBD has a higher affinity with the nitrocellulose membrane than the GFP control protein. We also tried to wash the proteins by using solutions with decreasing pH values. However, since high pH-steps were used in the pH-washes, all the proteins were gone when they were exposed to an acidic solution of pH3.
Coating an aerogel with mSA-GFP-CBD protein
With the production of the GFP fusion protein, we coated a hydrogel of 30 mm diameter and 3 mm thickness with 7.64 x 10^-10 mol of proteins. We used as a positive control a GFP that doesn’t contain the CBD domain. We then sent the coated hydrogels to the freeze drier to produce aerogels.
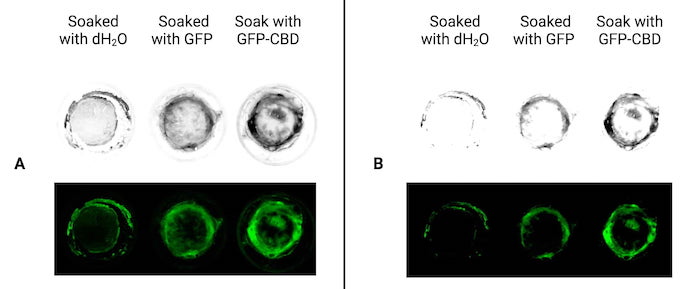
Figure 7 | Fluorescence images of the aerogels after the freeze-drying step produced from soaked hydrogels. (A) Aerogels produced from hydrogels previously soaked respectively with double-distilled water (negative control), GFP (positive control) and mSA-GFP-CBD (our protein of interest). (B) Aerogels produced from hydrogels previously soaked respectively with double-distilled water (negative control), GFP (positive control) and mSA-GFP-CBD (our protein of interest) but five days after the freeze-drying step.
- Analysis
As we can see on fig 6A, the aerogels were green under the fluorescence microscope which meant that the proteins bound the hydrogel and stayed on it during the freeze drying process. Even if we observed fluorescence for the three samples (water, control GFP and our recombinant GFP), the aerogel with mSA-GFP-CBD showed the most intense fluorescence. It showed that after the freeze-drying step, our mSA-GFP-CBD binds more to cellulose than usual GFP. The comparison of the fluorescence intensity between (fig 7.A) and (fig 7.B) showed that it has decreased with time. However, our mSA-GFP-CBD still showed the most intense fluorescence. This proves that our mSA-GFP-CBD bound tighter to cellulose thanks to its CBD domain compared to the usual GFP.
References
- Bauer, F. & Scheibel, T. (2012, 16 mai). Artificial Egg Stalks Made of a Recombinantly Produced Lacewing Silk Protein. Angewandte Chemie International Edition, 51(26), 6521‑6524. https://doi.org/10.1002/anie.201200591
- Bauer, F. (2013) Development of an artificial silk protein on the basis of a lacewing egg stalk protein. https://epub.uni-bayreuth.de/72/1/Diss.pdf
- Lim, K. H., Huang, H., Pralle, A. & Park, S. (2012, août 8). Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection. Biotechnology and Bioengineering, 110(1), 57‑67. https://doi.org/10.1002/bit.24605
| None |
