Part:BBa_K3171175
pTet-mCherry optimized in E. coli
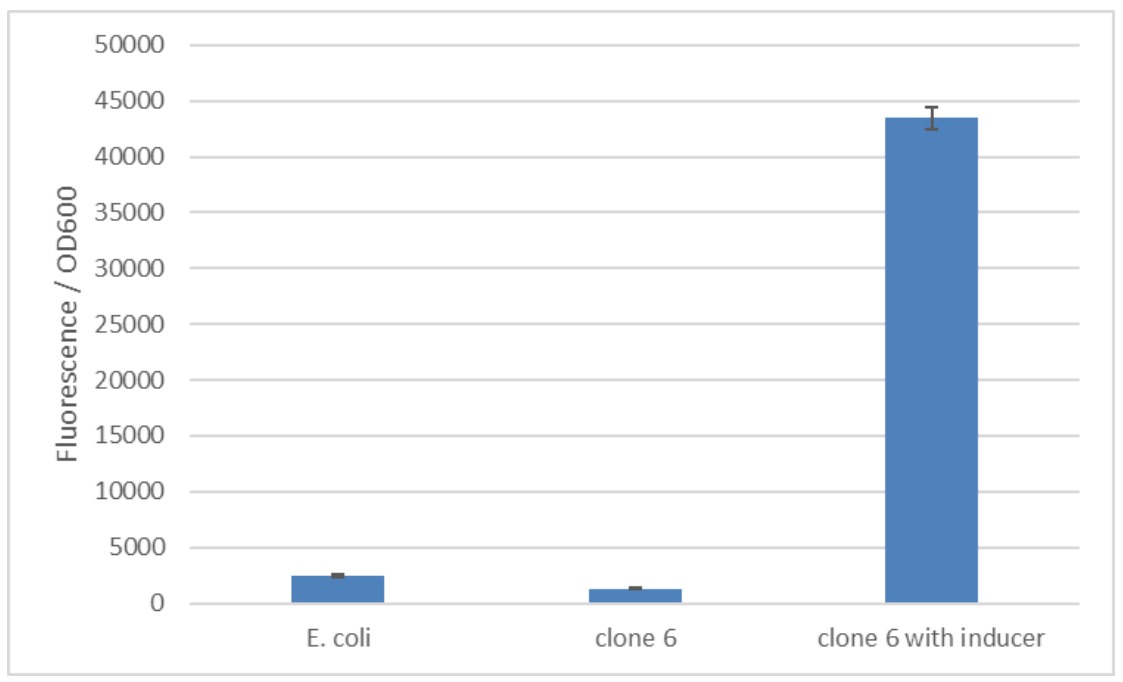
We have improved the pTET promoter's function and inducibility for use in E. coli employing a synthetic promoter library on pTet Part:BBa_R0040. The SPL was constructed using a PCR with randomized primers to yield arbitrary sequences in the promoter region. A total of 245 clones were screened to find a target with enhanced functionality. For the screening the cells were induced with 250 ng/mL anhydrotetracycline. The expression of mCherry was quantified based on the emitted fluorescence levels 8 hours after induction. The screening of the final 6 interesting variations revealed clone 6 as the sequence with increased inducibility (figure 1). The best version of the promoter was determined based on fold change in the fluorescence intensity and low basal level. We calculated a fold change of 34 comparing fluorescence in the induced and uninduced state! Additionally, this promoter also exhibited a low basal level of fluorescence that is comparable to background fluorescence of E. coli without any reporter (figure 2). The improved pTET promoter can be found at Part:BBa_K3171173.
Figure 1: Clone 6 exhibits the highest fluorescence fold change upon induction. Single clones from the synthetic promoter library were induced with 250 ng/ml anhydrotetracycline. The fluorescence fold change is calculated by dividing the fluorescence in the induced state by fluorescence in the absence of inducer. The used fluorescence reporter is mCherry.
Figure 2: The basal level of reporter expression is comparable to E. coli without the construct. Clone 6 from the synthetic promoter library was induced with 250 ng/ml anhydrotetracycline. The used fluorescence reporter is mCherry.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |