Part:BBa_K2151200
crtEBIY
This biobrick was created through standard biobrick assembly of K118014(RBS+crtE), K118006(RBS+crtB), K118005(RBS+crtI) and K118013(crtY). These genes are a part of the carotenoid biosynthesis pathway and together, this biobrick converts converts colourless farnesyl pyrophosphate to orange beta-carotene Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1974
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1510
Illegal NgoMIV site found at 1640
Illegal AgeI site found at 725 - 1000COMPATIBLE WITH RFC[1000]
Note added by Team Fudan 2022
As many researches indicate, the major problem of polycistronic vectors, which contain two or more target genes under one promoter, is the much lower expression of the downstream genes compared with that of the first gene next to the promoter[1]. Even though the original design here put crt genes order as how β-carotene was produced (crtE → crtB → crtI → crtY), the team failed to present data supporting their design works.
Instead of assembling CDSs sequentially, we construct a ribozyme-assisted polycistronic co-expression system (pRAP) by inserting ribozyme sequences between crtEBIY. In the pRAP system, the RNA sequences of hammerhead ribozyme conduct self-cleaving, and the polycistronic mRNA transcript is thus co-transcriptionally converted into individual mono-cistrons in vivo. Self-interaction of the polycistron can be avoid and each cistron can initiate translation with comparable efficiency. Besides, we can precisely manage this co-expression system by adjusting the RBS strength of individual mono-cistrons, as in BBa_K4162117.
Improved part by Team Fudan 2022
Our improved part is BBa_K4162117 and BBa_K4162118. We not only using ribozyme sequence flanking RBS+CDS, but also using a stronger RBS to drive crtE expression in biobrick crtEBIY.
Successful production of β-carotene in bacteria DH5α
Our BBa_K4162117 biobrick was created through overlapping PCR of BBa_K4162020(ribozyme+J6_RBS+crtY), BBa_K4162010(ribozyme+T7_RBS+crtE), BBa_K4162013(ribozyme+T7_RBS+crtB) and BBa_K4162016(ribozyme+T7_RBS+crtI). We transfected BBa_K4162117 into E. coli DH5α to build single-cell factory for β-carotene production. Coding sequences of crtYEBI are separated by ribozyme sequences. In this part, the RBS of crtEBI has equal intensity while the RBS of crtY is significantly weaker than the others. Because crtY catalyzes the last step of the carotenoid reaction chain, we guess the concentration of substrate catalyzed by this enzyme is significantly lower than for the first three steps of the reaction. To avoid the problem of flux imbalance in biosynthesis as well as to reduce unnecessary metabolic stress on cells, we intentionally weakened the RBS intensity of crtY, and created BBa_K4162117. Please notice, our BBa_K4162118 places crt genes in the same order as BBa_K2151200, but not BBa_K4162117.
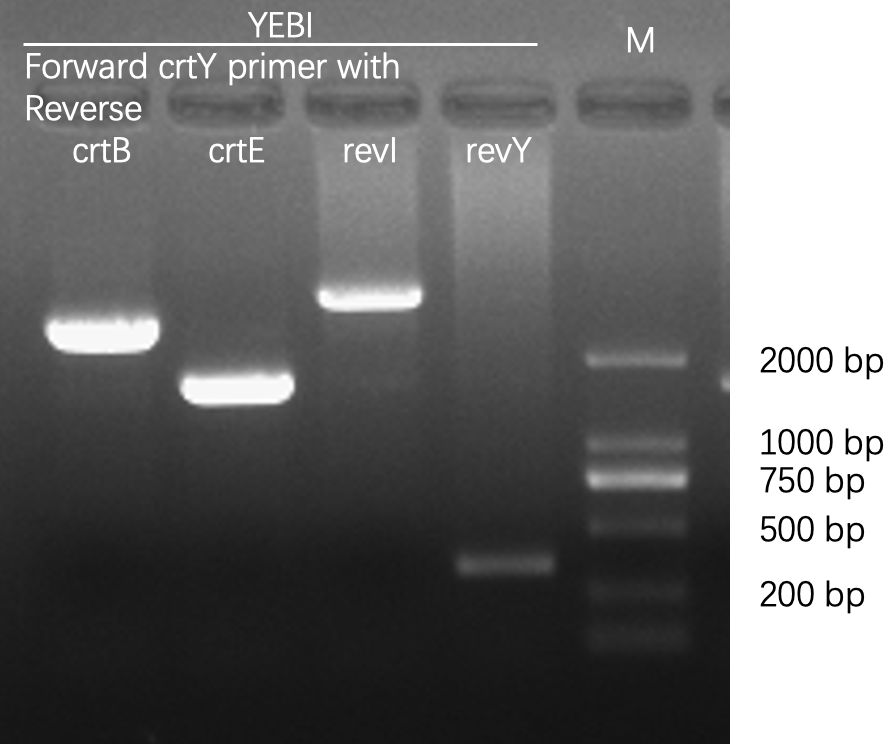
Characterization of crtYEBI
Agarose gel electrophoresis
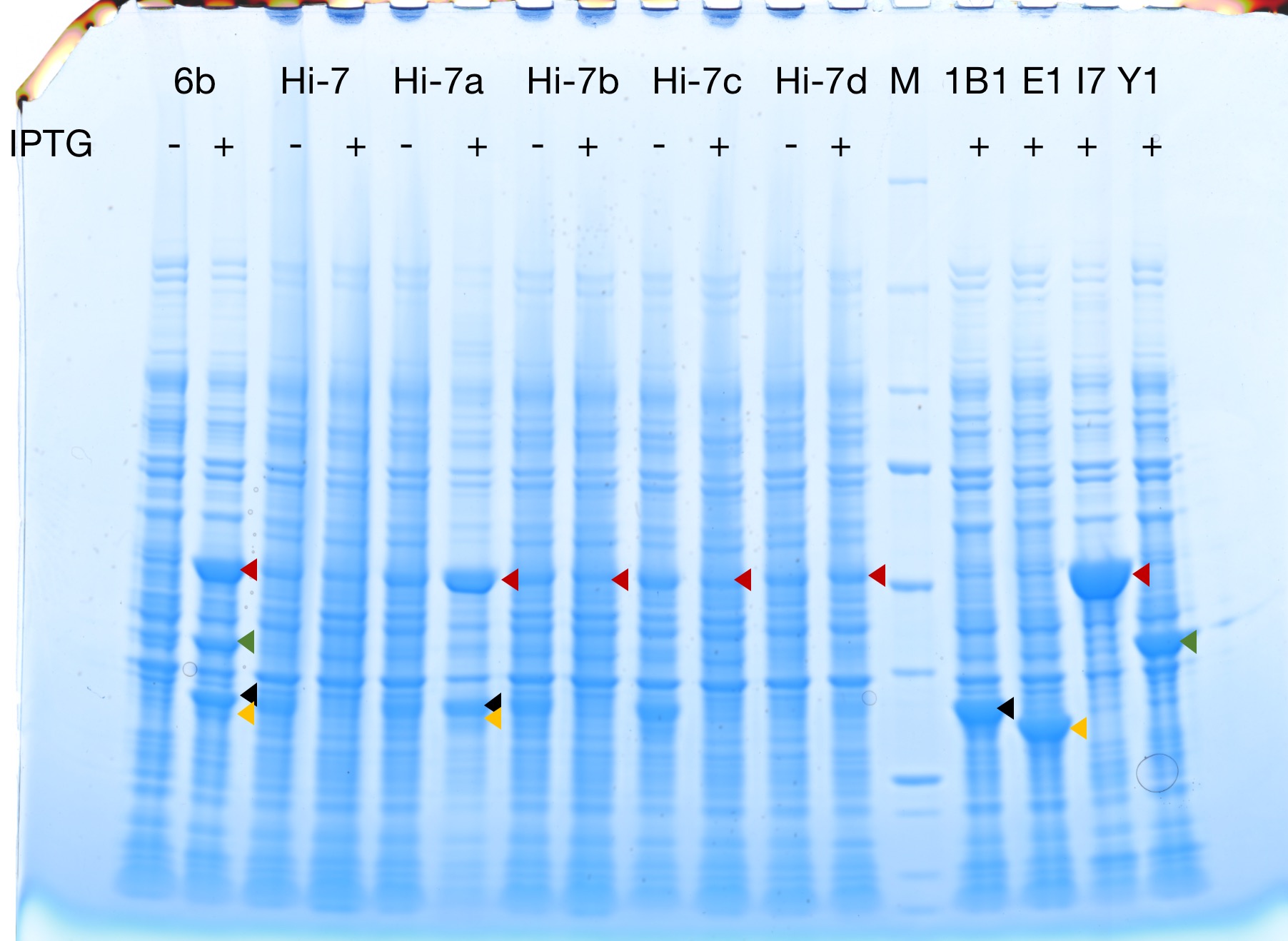
Successful protein expression
Produce β-carotene
Figures 2 to 5 show that E. coli transfected with BBa_K4162117 successfully expressed the target enzyme and yielded β-carotene. In Figure 5, it can be seen that module YEBI corresponds to a darker orange color of the post-centrifugation precipitation compared to module YBEI (BBa_K4162119), characterizing the superior carotenoid yielding ability of module YEBI.
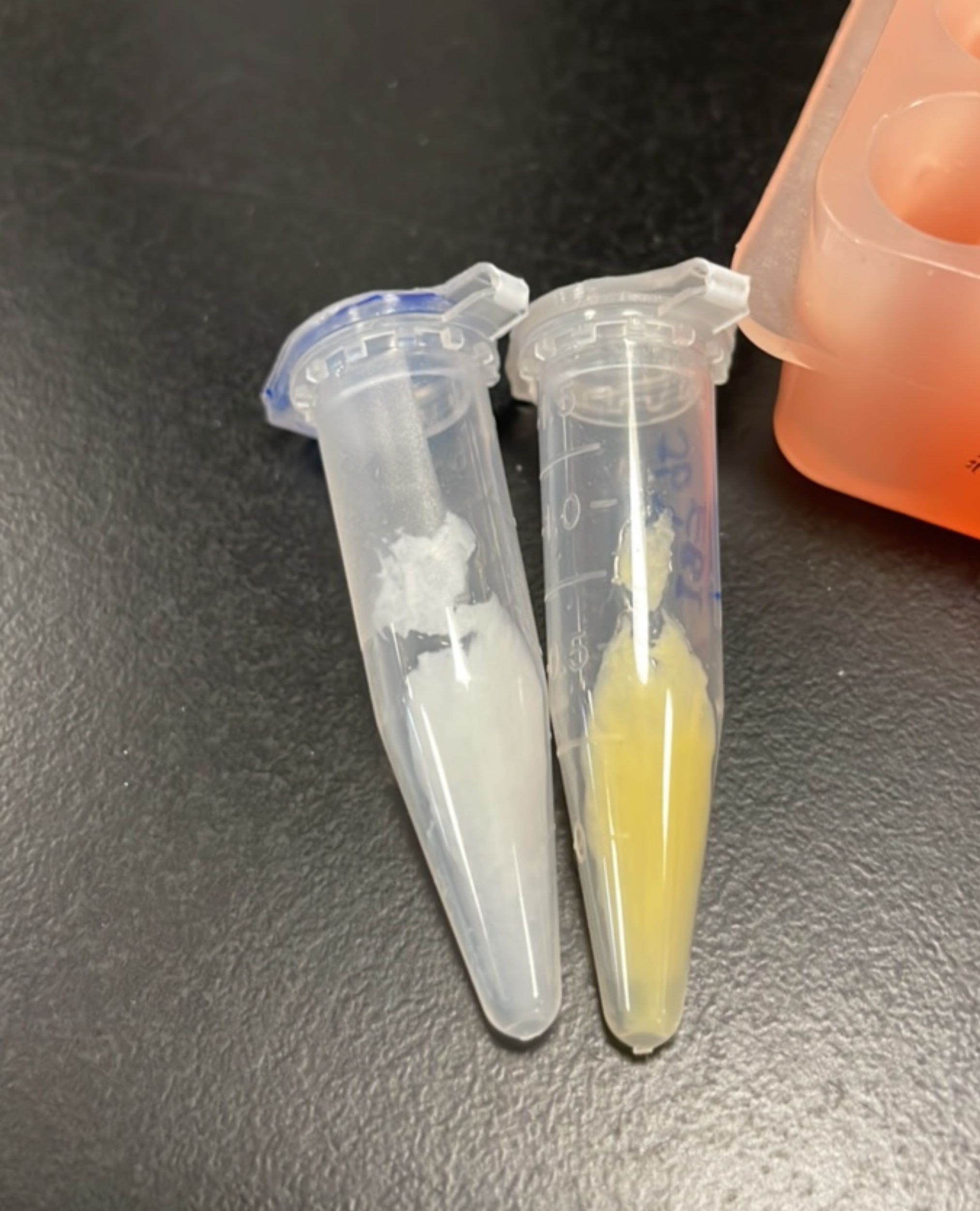

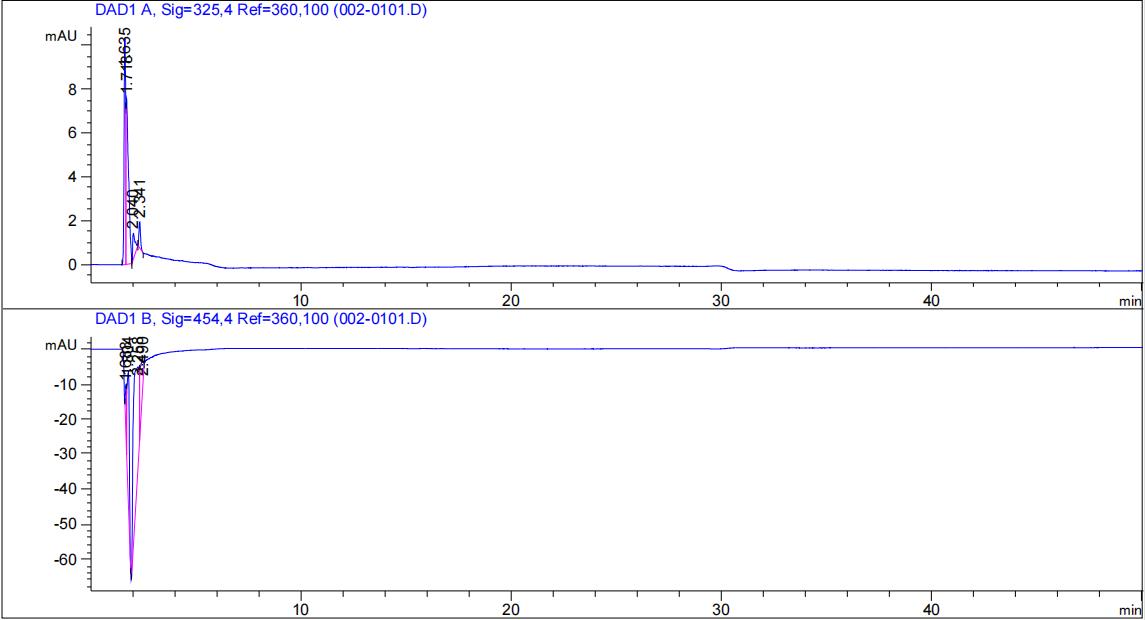
HPLC validation
Agilent liquid chromatograph (HPLC-DAD); column C18 (250mm); column temperature 30°C; mobile phase methanol:water = 96:4; flow rate 0.8 mL/min; detection wavelength 325 nm and 454 nm.

References
- ↑ Kim, K. J., Kim, H. E., Lee, K. H., Han, W., Yi, M. J., Jeong, J., & Oh, B. H. (2004). Two-promoter vector is highly efficient for overproduction of protein complexes. Protein science : a publication of the Protein Society, 13(6), 1698–1703.
| None |