Part:BBa_K1982008
tCas9-CIBN (Eukaryotic LACE system)
| RBS-tCas9-CIBN-HA | |
|---|---|
| Function | gene activation |
| Use in | Eukaryotic cells |
| RFC standard | RFC 10 |
| Backbone | pSB1C3 |
| Submitted by | [http://2016.igem.org/Team:NEU-China NEU-China 2016] |
The NEU-China iGEM team 2016 designed a fusion protein consisting of tCas9 and CIBN for sequence-specific transactivation of a desired target locus (more information). We used our double truncated tCas9 (BBa_K1982001) impaired in its cleavage activity and fused it to the CIBN (BBa_K1982003). An prokaryotic RBS sequence from the Community collection(BBa_B0034) fused to the beginning of tCas9-CIBN. For detection of expression the fusion protein was tagged with a HA-epitope coding sequence (BBa_K1150016).

|
| Figure 1: Construct design. tCas9 was fused to CIBN. The resulting fusion construct was flanked by RBS sequences and tagged by a HA epitope. The pBad/araC promoter and rrnB T1 terminator were chosen to control gene expression. |
Optogenetic systems enable precise spatial and temporal control of cell behavior. A light-activated CRISPR/Cas9 effector (LACE) system that induces transcription of endogenous genes in the presence of blue light.This was accomplished by fusing the light-inducible heterodimerizing proteins CRY2 and CIB1 to a transactivation domain and the catalytically inactive tCas9, respectively. The versatile LACE system can be easily directed to new DNA sequences for the dynamic regulation of endogenous genes[1].
we fused the light-inducible heterodimerizering proteins CRY2 and CIB1 from Arabidopsis thaliana to the VP64 transactivation domain and C-terminus of tCas9.
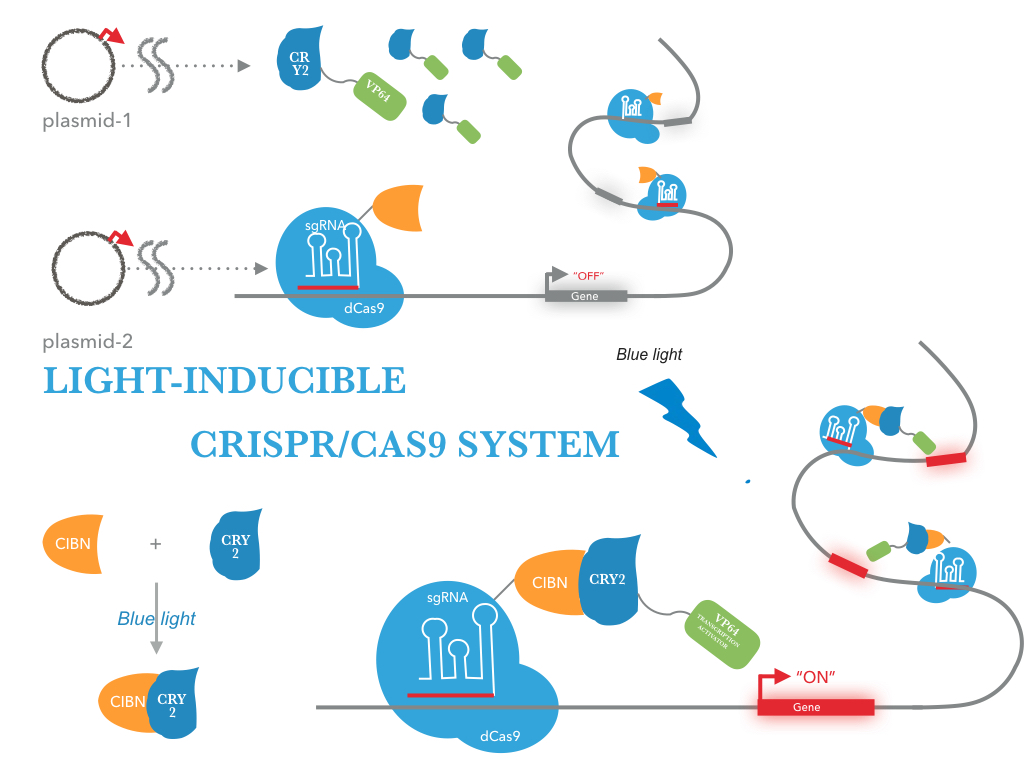
|
| Figure 2: Figure 1 illustrates the detailed design of LACE device |
Usage and Biology
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 10: (underlined part encodes the protein) ATGGACAAG ... GATTACGCTTAATAA ORF from nucleotide position 1 to 4710 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| Alignments (obtained from PredictProtein.org) There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
| There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Proof of function
Verification of the suppression efficiency of gRNA
With different target loci have been tested by the usage of a GFP reporter plasmid(pCold-1) with a CSPA promotor. The target sites can be determined by directing the gRNA consisting of 20 bp length against the desired sequence of interest. F1 gRNA with target sites at different distances to the promotor regions proved successfully as potential activation sites (see Table 1 and Figure 3).
| Name | Binding Site | Distance to promoter | Sequence | Position |
| F1 | CSPA promoter | 15 | TGCATCACCCGCCAATGCG | sense sequences |
| F2 | non-coding | 68 | GCCGCCGCAAGGAATGGTG | sense sequences |
| R1 | CSPA promoter | 43 | ATTAATCATAAATATGAAA | antisense sequences |
| R2 | non-coding | 94 | CATCATCCAACTCCGGCAAC | antisense sequences |
Table 1: Overview of the tested gRNAs with different binding sites on the GFP pCold-1 plasmid.

|
| Figure 3: Position of the target loci on the GFP pCold-1 plasmid. |
To ensure the suppression efficiency of the gRNA, four gRNA sequences targeting different sites of CSPA promoter were designed and transfected into the E. coli strains BL21. Efficient suppression of CSPA promoter in strains BL21 was observed. GFP levels in GFP transgenic strains decrease after inserting a fragment that expresses CSPA promoter gRNA. And the sequence with the best suppression effect was selected for further study.

|

|
| Figure 4: Results of the GFP-influence under the CSPA promotor only using different gRNAs targeted to CSPA promoter in BL21.. 1-8: transfected with different GFP-gRNA plasmid (CSPA promoter) 9:Control group transfected with GFP plasmid,~27 kDa. Excitation wavelength: 488 nm, Emission wavelength: 509nm. Stationary cultures of BL21 was subcultured into fresh media and growth for 8 hours (1 3 5 7 9) or 16 hours (2 4 6 8) at 30°C. 30ng of protein with total volume of 30ul (protein sample + dissociation buffer). |
Silencing capability validation
We next evaluated the effect of tCas9-cibn on suppressing CSPA promoter. GFP expression levels were assayed in strains BL21 after co-transformation with tCas9-cibn and gRNA.

|

|
| Figure 5: Silencing capability of tCas9-CIBN with gRNA using different gRNAs targeted to CSPA promoter in BL21. 1-8: transfected with different GFP-gRNA plasmid (CSPA promoter) and tCas9-CIBN plasmid(pBAD promoter) 9 10:Control groups transfected with GFP plasmid and tCas9-CIBN plasmid(pBAD promoter), ~27 kDa. Excitation wavelength: 488 nm, Emission wavelength: 509nm. Stationary cultures of BL21 was subcultured into fresh media and growth for 8 hours (1 3 5 7 9) or 16 hours (2 4 6 8 10) using 15mM L-arabinose at 30°C. 30ng of protein with total volume of 30ul (protein sample + dissociation buffer). |
Compared with control groups, green fluorescence intensity and mRNA levels were dramatically reduced in groups treated with gRNA and tCas9-cibn. These results suggest that gRNA can specifically guide tCas9 to target upstream of CSPA promoter, thereby to inhibit CSPA promoter to reduce GFP expression levels.
Activation of a fluorescence reporter
Spatially controlled activation of gene expression was achieved in strains co-transfected with the LACE system, a reporter vector containing a gRNA target sequence upstream of CSPA promoter and the eGFP gene. Strains transfected with light-activated CRISPR/Cas9 effector (LACE) and incubated in the dark did not show a significant difference in eGFP levels compared to control groups transfected with empty plasmid. Strains containing the LACE system and gRNA exhibited significantly brighter eGFP fluorescence intensity when illuminated compared to when incubated in the dark.Activation of the eGFP reporter in strains transfected with the gRNA and LACE constructs, the gRNA and tCas9-VP64 expression plasmid or an empty plasmid as a negative control was quantified after 24 hours of illumination or incubation in the dark.

|
| Figure 6: Activation capability of LACE system using gRNA-F1 plasmid and tCas9-CIBN plasmid in BL21. Excitation wavelength: 488 nm, Emission wavelength: 509nm. Stationary cultures of BL21 was subcultured into fresh media and growth for 8 hours using 15mM L-arabinose at 30°C. |
Proof of expression
Stationary cultures of BL21 pBAD was subcultured into fresh media and induced for 4 hours or 16 hours using different concentrations L-arabinose. Subsequent purification of protein from the cell-free supernatant and visualization using SDS-PAGE confirms that proteins of the expected size are present in the supernatant and hence most likely successfully secreted by the engineered bacterial strains.


|
| Figure 7: Western blot analysis of tCas9-CIBN protein levels. 1 3 5 7 9 11:Control groups transfected with empty plasmid; 2 4 6 8 10 12: transfected with tCas9-CIBN plasmid (pBAD promoter), ~180 kDa. Stationary cultures of BL21 was subcultured into fresh media and growth for 4 hours or 16 hours using different concentrations L-arabinose. 30ng of protein with total volume of 30ul (protein sample + dissociation buffer). |
Usage and Biology
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2758
Illegal NgoMIV site found at 3667
Illegal NgoMIV site found at 4231 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 3786
Illegal SapI.rc site found at 1177
Illegal SapI.rc site found at 1419
[1] Westra E.R., Swarts D.C., Staals R.H., Jore M.M., Brouns S.J., van der Oost J. (2012). The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet. 46, 311-39
[2] Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339(6121), 823-6
[3] Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 31(3), 233-9
[4] Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., Zhang, F. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339 (6121), 819-23
[5] Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5), 1173-83
[6] Lauren R. Polstein and Charles A. Gersbach. (2015). A light-inducible CRISPR/Cas9 system for control of endogenous gene activation. Nat Chem Biol 11(3): 198–200
//function/crispr/cas9
| direction | Forward |
