Part:BBa_J428353
pJUMP28-1A KanR Type IIS Level 1 vector. Origin pUC (high copy number)
Contents
Characterisation Studies
Cambridge 2022 JUMP DV Characterisation
- Summary
- Verdict
- Experience/Detailed Protocol: https://parts.igem.org/wiki/index.php?title=Part:BBa_J428351:Experience
DNA Sequence and Features
Assembly Compatibility:- 10INCOMPATIBLE WITH RFC[10]Illegal prefix found at 2492
Illegal suffix found at 16 - 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 2492
Illegal SpeI site found at 17
Illegal PstI site found at 31
Illegal NotI site found at 24
Illegal NotI site found at 2498 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 2492 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 2492
Illegal suffix found at 17 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 2492
Plasmid lacks a suffix.
Illegal XbaI site found at 2507
Illegal SpeI site found at 17
Illegal PstI site found at 31
Illegal NgoMIV site found at 1390 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal SapI.rc site found at 2188
- 10
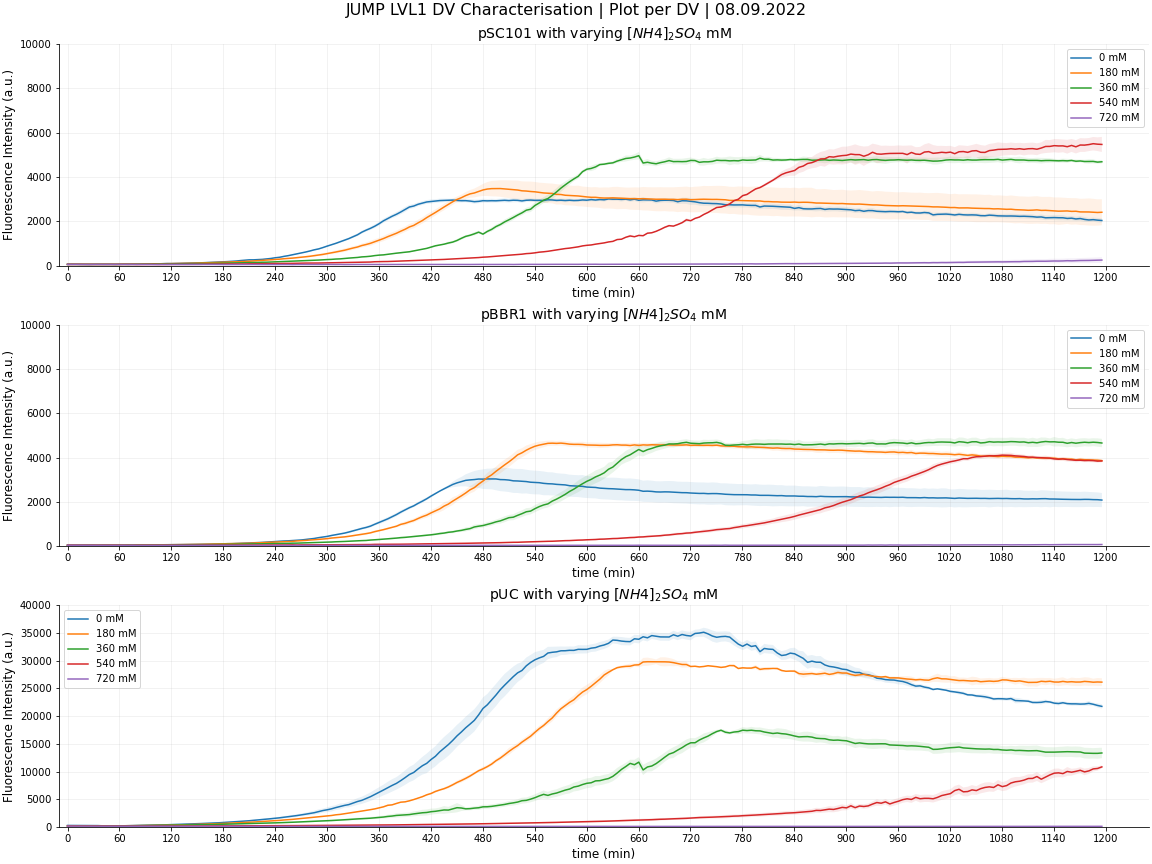
As part of our 'Bronze - Contribution' criteria, Cambridge 2022 characterised 3 JUMP DV plasmids, from the 2022 distribution kit plate 1, in DH5α E. coli against increasing concentrations of ammonium sulphate. The DVs were of varying copy number: pSC101 (low copy no.), pBBR1 (low-med. copy no.) and pUC (high copy no.) to investigate the effect of ammonium on bacterial growth and expression with varying copy number burden.
DV pSC101 (low copy no.): Ammonium sulphate enhanced fluorescence intensity up to 540mM. Despite the fluorescence rate decrease, the maximum fluorescence increased as we increased ammonium sulphate concentration to 540mM with the cells reaching their limit at 720mM. Therefore, up to a maximum of 540mM of ammonium sulphate can be added to the growth media of cells containing pSC101 DV in order to enhance the expression of the protein of interest (in place of our sfGFP).
DV pBBR1 (low-med copy no.): Ammonium sulphate enhanced fluorescence intensity up to 360mM. Despite the fluorescence rate decrease, the maximum fluorescence increased as we increased ammonium sulphate concentration to 360mM with the cells having lower expression at 540mM and finally reaching their limit at 720mM. Therefore, up to a maximum of 360mM of ammonium sulphate can be added to the growth media of cells containing pBBR1 DV in order to enhance the expression of the protein of interest (in place of our sfGFP).
DV pUC (high copy no.): For high copy number plasmids, we can report that any ammonium concentration is detrimental to the cells expression. The greatest fluorescence was recorded with 0mM of ammonium sulphate, with decreasing expression as it was increased.
//collections/jump/level1
//plasmidbackbone/assembly/typeiis
//plasmidbackbone/copynumber/high
| resistance | kanamycin |