Part:BBa_R0084
Promoter (OmpR, positive)
Positively regulated, OmpR-controlled promoter. This promoter is taken from the upstream region of ompF. Phosphorylated OmpR binds to the three operator sites and activates transcription.
Usage and Biology
In nature, this promoter is upstream of the ompF porin gene and is both activating and repressing. The regulation of ompF is determined by the EnvZ-OmpR osmosensing machinery. EnvZ phosphorylates OmpR to OmpR-P. At low osmolarity, EnvZ has a low activity, producing a small concentration of OmpR-P. This OmpR-P binds to high-affinity sites on ompF, activating transcription. At high osmolarity, however, EnvZ is more active, creating a high OmpR-P concentration. OmpR-P then binds to the low-affinity sites on ompF, repressing transcription. See Notes section for further discussion of negative regulation.
Contribution: HKUST 2021
Summary
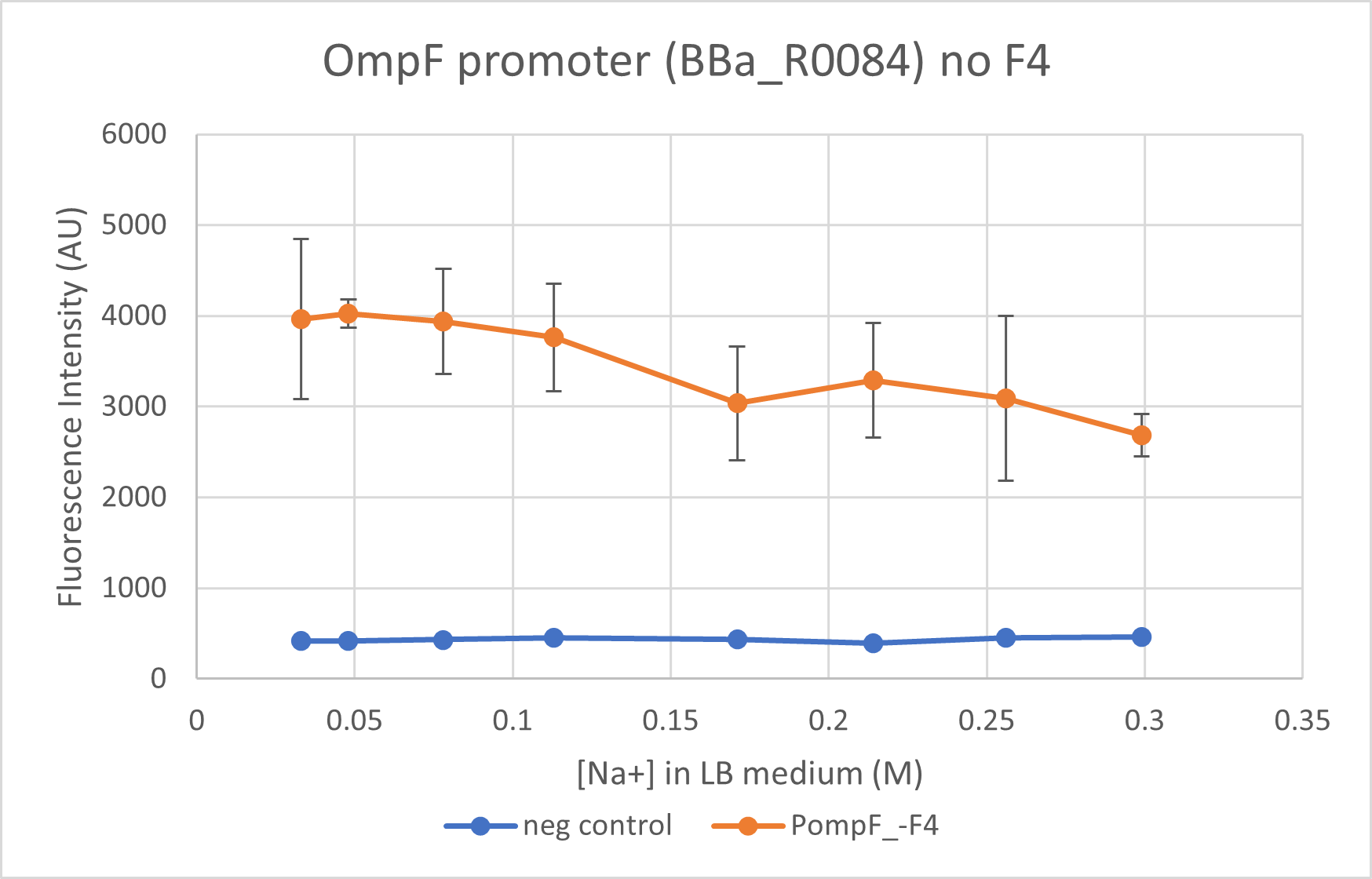
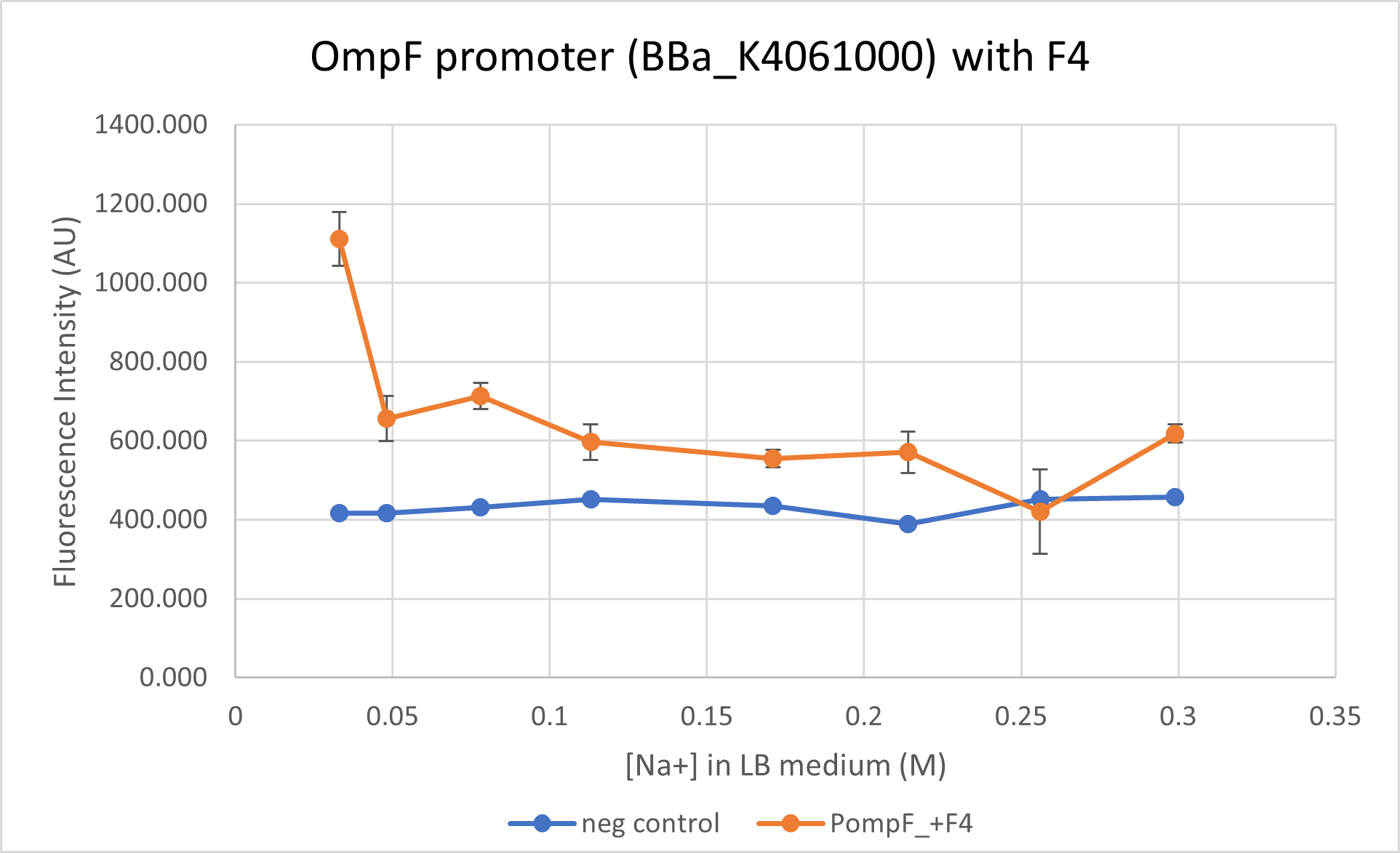
This is characterisation as well as proposed improvement for the part. We characterised the OmpF promoter with (BBa_K4061000) and without its negative regulator (BBa_R0084) region. This part does not contain the negative regulator domain F4 and we compare its activity to BBa_K4061000 which contains this domain. We observe that the presence of the negative regulator allows a better control of the promoter and allows a "switch-like" response for an increasing osmolarity.
Experiments
We got the complete sequence of OmpF promoter (with negative regulator) from E. coli K12 strain genome. We ordered the whole sequence in tandem with the mRFP1 reporter via IDT. This gave us the part BBa_K4061000. In order to analyse the action of negative regulator we PCRed out the fragment right before the positive regulators in order to eliminate the negative regulator. The resulting sequence is BBa_R0084 OmpF promoter with mRFP1 reporter. Cells transformed with these two constructs were cultured in LB media with varying NaCl concentrations and the fluorescence intensities were compared at different media concentrations - final Na+ concentrations of 0.033M, 0.048M, 0.078M, 0.131M. 0.171M, 0.214M, 0.256M and finally 0.299M. Concentrations below 0.171M are considered as "low osmolarity" from the ideal LB medium recipe.
Results and Discussion
We observe a steeper decline close to wild-type level for mRFP1 regulated under OmpF promoter with the negative regulator. The OmpF promoter without the negative regulator reduces expression of reporter only 1.6 times at a high osmolarity while the promoter with negative regulator reduced expression to 2 times. BBa_R0084 (without F4) regulated mRFP1, however, has a much higher fluorescence intensity as compared to the reporter regulated under BBa_K4061000 (with F4).
Note: Data from the two constructs, despite being taken concurrently, is not plotted on the same graph due to the difference in the intensities. Separating the graphs allow better visualisation of the trend in the promoter activity.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
//direction/forward
//chassis/prokaryote/ecoli
//promoter
//regulation/positive
//classic/regulatory/uncategorized
| biology | |
| direction | Forward |
| negative_regulators | |
| positive_regulators | 1 |

 1 Registry Star
1 Registry Star