Part:BBa_R0082
Promoter (OmpR, positive)
Positively regulated, OmpR-controlled promoter. This promoter is taken from the upstream region of ompC. Phosphorylated OmpR binds to the three operator sites and activates transcription.
Usage and Biology
In nature, this promoter is upstream of the ompC porin gene. The regulation of ompC is determined by the EnvZ-OmpR osmosensing machinery. EnvZ phosphorylates OmpR to OmpR-P. At high osmolarity, EnvZ is more active, creating more OmpR-P. OmpR-P then binds to the low-affinity OmpR operator sites upstream of ompC.
Contribution: Hamburg 2016
Group: Hamburg / Authors: Kai Pohl, Daniel Wedemeyer
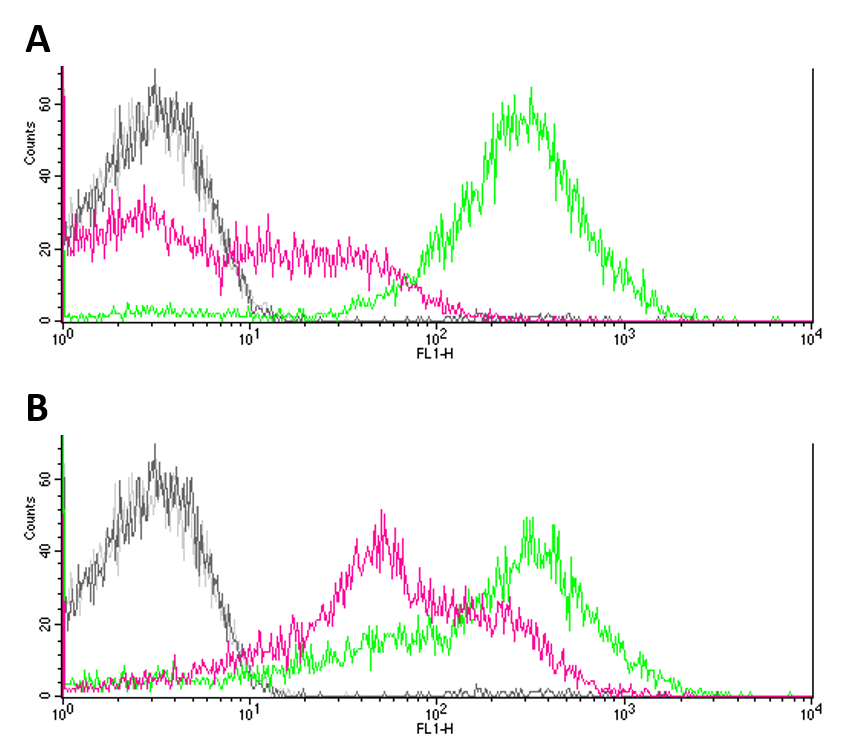
Summary: We analysed the noise produced by BBa_R0082 by flow cytometry using a ΔEnvZ strain.
In two approaches, we ligated BBa_R0082 (OmpR, positive promoter) with BBa_E0240 (GFP) and BBa_E0430 (eYFP) into pSB1C3 transformed both ΔEnvZ (JW3367-3) and wt (DH5a) E. coli strains with them. Constructs were validated by sequencing. We submitted the constructs as: BBa_K1909013 (Omp/GFP) and BBa_K1909014 (Omp/eYFP) to the registry.
Experiments
To determine the noise of the OmpR promoter, we measured the expression of our Fluorescence reporters by flow cytometry, comparing the ΔEnvZ (JW3367-3) to wt (DH5a) E. coli strain. Furthermore, we used JW3367-3 and DH5a strains without our reporter system as negative controls. Cells were incubated in LB medium to OD600 = 0.2, washed using 100 mM MgCl2 and diluted 1:100 prior to measuring.
Results
Negative controls (JW3367-3 and DH5a without reporter systems) showed only few to no fluorescence (Fig.1 A, B). GFP expression (Fig. 1 A) proved to be significantly higher in DH5a (green) cells as compared to EnvZ deficient JW3367-3 cells (pink). Although not as pronounced, this difference was also observable for eYFP expression in the respective cell lines (fig. 1 B). Differences in the expression of fluorescence reporters were confirmed by KS tests (GFP: p <= 0.001, eYFP: p <= 0.001).
Leaky Expression by the OmpR-Regulated Promoter on Different Vectors
(Characterized by SDU-Denmark)
Leaky expression by the OmpR-regulated promoter is reduced when cloned into a low copy vector compared to a high copy vector.
The expression properties of the OmpR-regulated promoter were investigated using a reporter system containing RFP under control of the OmpR-regulated promoter, BBa_M30011, was cloned into E. coli strain SØ928 ΔompR, lacking the OmpR transcription factor, on a high copy vector. By using a ΔompR strain, the background generated by stimulation of the intrinsic OmpR system is removed, and the strain functions as a negative control.
RFP expression was assessed by fluorescence microscopy using an Olympus IX83 with a photometrics prime camera, with exposure time for RFP at 200 ms.
Assessing the RFP expression by fluorescence microscopy, it was discovered that the OmpR-regulated promoter mediated gene expression even in the absence of its transcription factor, see Figure 1. This observation was confirmed by going through the literature [1].

On the basis of this finding, controlled gene expression by the OmpR-regulated promoter required a low copy plasmid or insertion into the chromosome. Protein expression of RFP in pSB1C3 with a copy number of 100-300 plasmids per cell, and pSB3K3 with a copy number of 10-12 plasmids per cell, was studied by flow cytometry. As for the determination of noise levels in the weak, BBa_J23114, and strong BBa_J23102constitutive promoters, the experiment was carried out in both LB medium and M9 minimal medium, the latter supplemented with 0.2% glycerol. In the LB medium, selection was carried out by the addition of 30 µg/mL chloramphenicol, 30 µg/mL kanamycin, or 50 µg/mL ampicillin, depending on the resistance, and for M9 minimal medium, the concentrations used were 60 µg/mL chloramphenicol, 60 µg/mL kanamycin, and 100 µg/mL ampicillin. Excitation of RFP was at 561 nm, and emission was measured around 580 nm. Expression levels in both E. coli MG1655 and E. coli MG1655 ΔompR were studied to determine the baseline of the leaky expression not influenced by intrinsic pathways including the OmpR transcription factor.

Fluorescence levels in the two different media display similar behavior, as seen in Figure 2. The main difference observed, was that the decrease in fluorescence over time was faster in LB medium than in M9 minimal medium, in concordance with the observations made in previous experiments. On a general level, the data revealed, that MG1655 cloned with the POmpR-RFP reporter system on the high copy vector exhibited a fluorescence level, equivalent to that mediated by the strong constitutive promoter. On the low copy vector, the POmpR-RFP reporter system yielded a fluorescence level comparable to the gene expression mediated by the weak constitutive promoter. On the other hand, expression levels in the MG1655 ΔompR strain were markedly reduced compared to MG1655, indicating that pathways including the transcription factor OmpR interfere with RFP expression under these conditions. Again, the fluorescence levels observed for the POmpR-RFP reporter system on the low copy vector were distinctly lower than for the high copy vector.
All things considered, the OmpR-regulated promoter was found to exhibit leaky expression comparable to the expression levels mediated by the constitutive promoters. When cloned into a low copy vector, the leaky expression was reduced prominently. Thus, to obtain proper regulation of gene expression by the OmpR-dependent promoter, a low copy vector is required.
Part:BBa_R0082 was first submitted by the Antiquity 2004 team, and additional characterizations were performed by the Hamburg 2016 team. For the 2019 edition, our team will provide additional experimental data related to the influence of this part in the growth kinetics of a wild type NEB-5 alpha strain. For this, we will use a construct comprised of ompC+GFP cloned into the pSB1C3 vector to transform NEB-5 alpha cells. We will then grow our transformants in media with varying concentrations of NaCl and sucrose to evaluate their growth kinetics at high osmolarity using a standard growth curve assay.
Petri dishes observed in UV transilluminator from NEB 5-alpha E. coli cells[pOmpC-GFP].
Miniprep plasmid extraction from pOmpC-GFP in transformed NEB 5-alpha Competent E. coli cells by QIAprep® Spin Miniprep Kit. 10kb: Quick-Load Purple 2-log DNA Ladder (10kb). M(1-3): Plasmid extraction triplicate.
Culture conditions and determination of cell growth and fluorescence
E. coli was grown in Luria-Bertani broth at 37°C with vigorous shaking. When necessary, ampicillin (Amp) (Sigma-Aldrich, St Louis, MO, USA) was added to the culture medium (100 µg/mL) Plate media were prepared by adding agar to liquid broth at a final concentration of 1.5%. Osmotic pressure conditions studied included NaCl (1, 5, 10, 20 and 40 g/L, and without NaCl), and dextrose (5 and 10%).
For the assessment of expression of GFP fluorescence during growth, the microtiter plate assay system Varioskan Flash (Thermo Fisher Scientific, Waltham, MA, USA) was used. All measurements were conducted in sterile 96-well bottom microplates (Nunc, Rochester, NY, USA) with a final assay volume of 300 µl/well. Overnight cultures were diluted into fresh LB medium before growth with different appropriate experimental additives (NaCl or dextrose). The microplates were incubated for 24 h at 37°C. Measurements were made at 1 h intervals. During cultivation, the Varioskan Flash provided quantitative online data of (i) cell density via measuring OD600 and (ii) in vivo GFP expression: GFP fluorescence was monitored at 511 nm upon excitation at 488 nm.
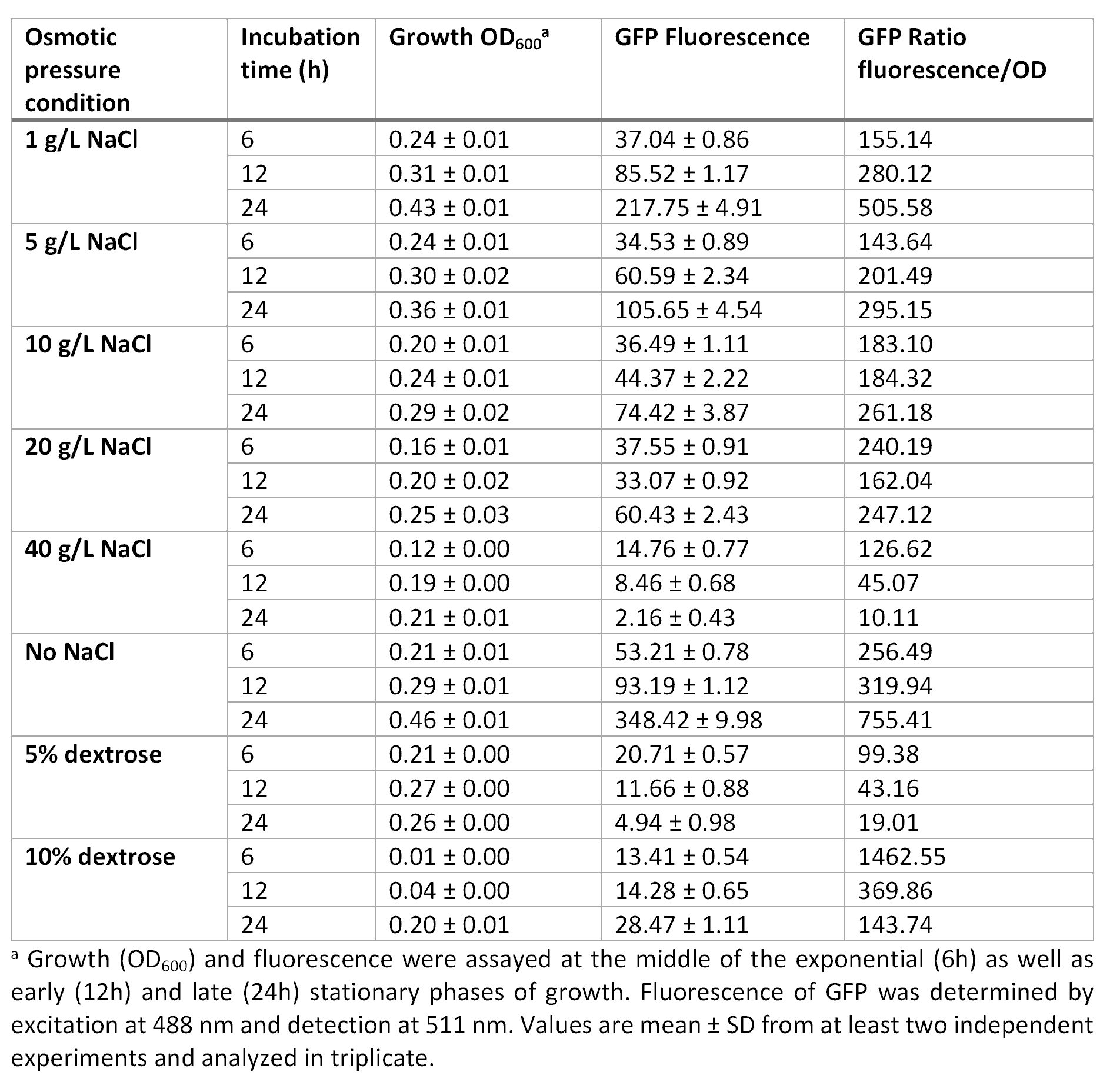
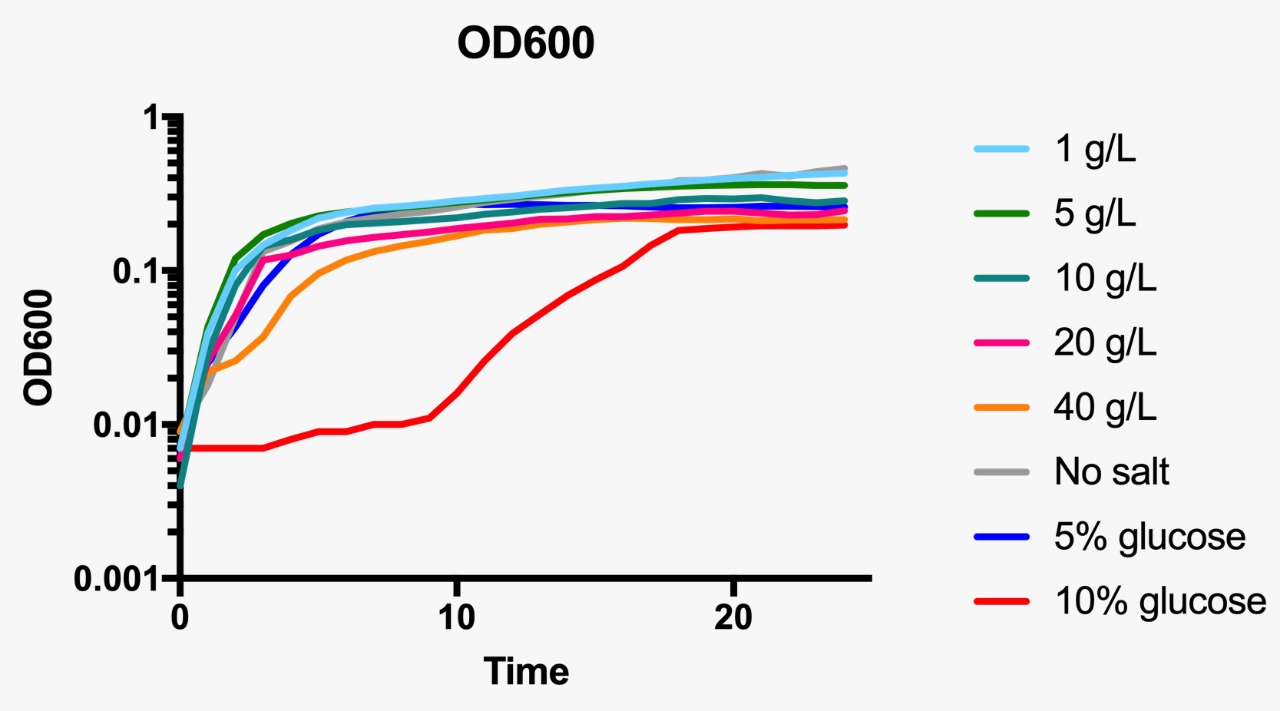
Growth curve at OD600 at different osmotic pressure conditions.
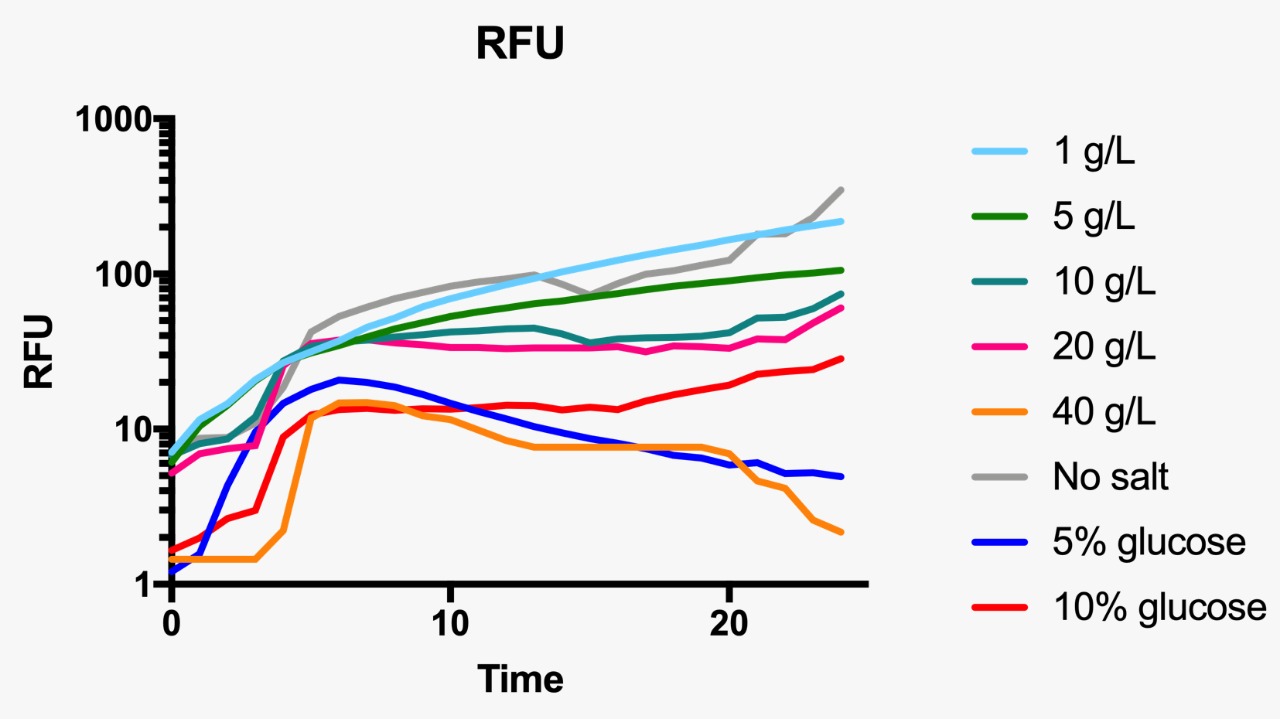
Fluorescence curve of GFP at 488/511 nm at different osmotic pressure conditions.
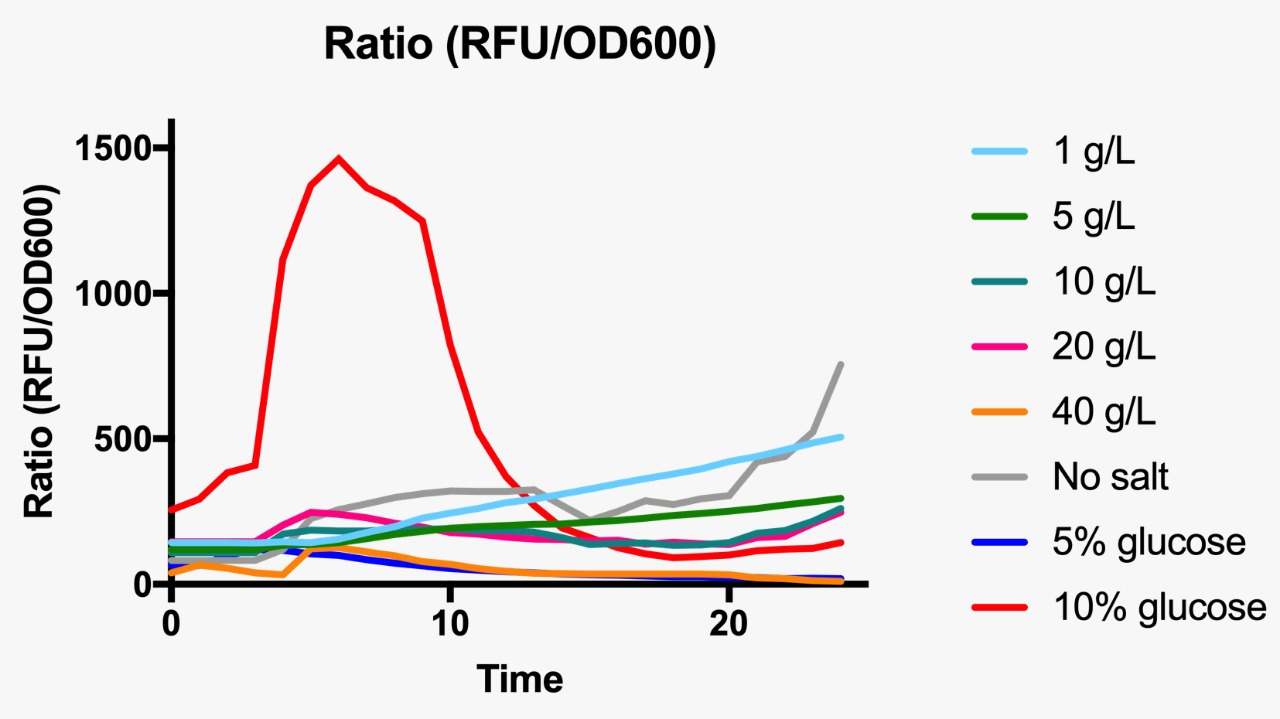
Ratio based on GFP fluorescence per optical density unit at different osmotic pressure conditions.
Contribution: HKUST 2021
Summary
We analysed the activity and response of OmpC promoter (BBa_R0082) to an increasing medium osmolarity. We utilised this promoter to regulate a reporter protein and observed the intensity of fluorescence as a measure of promoter activity. eGFP fluorescence intensity increased 2 fold with an increasing osmolarity which supports the hypothesis that OmpC promoter is activated at high osmolarity.
Experiments
We prepared LB growth media for E. coli BL 21 strain with varying amounts of NaCl- with final Na+ concentrations of 0.033M, 0.048M, 0.078M, 0.131M. 0.171M, 0.214M, 0.256M and finally 0.299M. Cells containing the composite part BBa_K4061070 which has OmpC regulated eGFP were cultured in the above media for 16 hours. We measured fluorescence intensity from the culture for all different concentrations along with negative-plasmid control, wild type BL 21 cells. The samples were excited at 488 nm and emission was read at 507 nm. In order to correlate the external medium concentration to intracellular osmolarity, we lysed cultured cells and measured the osmolarity of the supernatant using an osmometer. We found an exponential relationship between extracellular medium osmolarity and intracellular osmolarity. For clarity sake, we will still present data in terms of external medium concentration.
Results and Discussion
With increasing medium osmolarity, as a result of increasing medium Na+ concentration, the eGFP intensity is seen to increase 2 fold. The intensity increases from 10,000 AU to 20,000 AU. A negative plasmid control with WT BL21 cells is also measured for fluorescence intensity. Therefore, we can conclude that PompC is favourably activated at higher osmolarities.
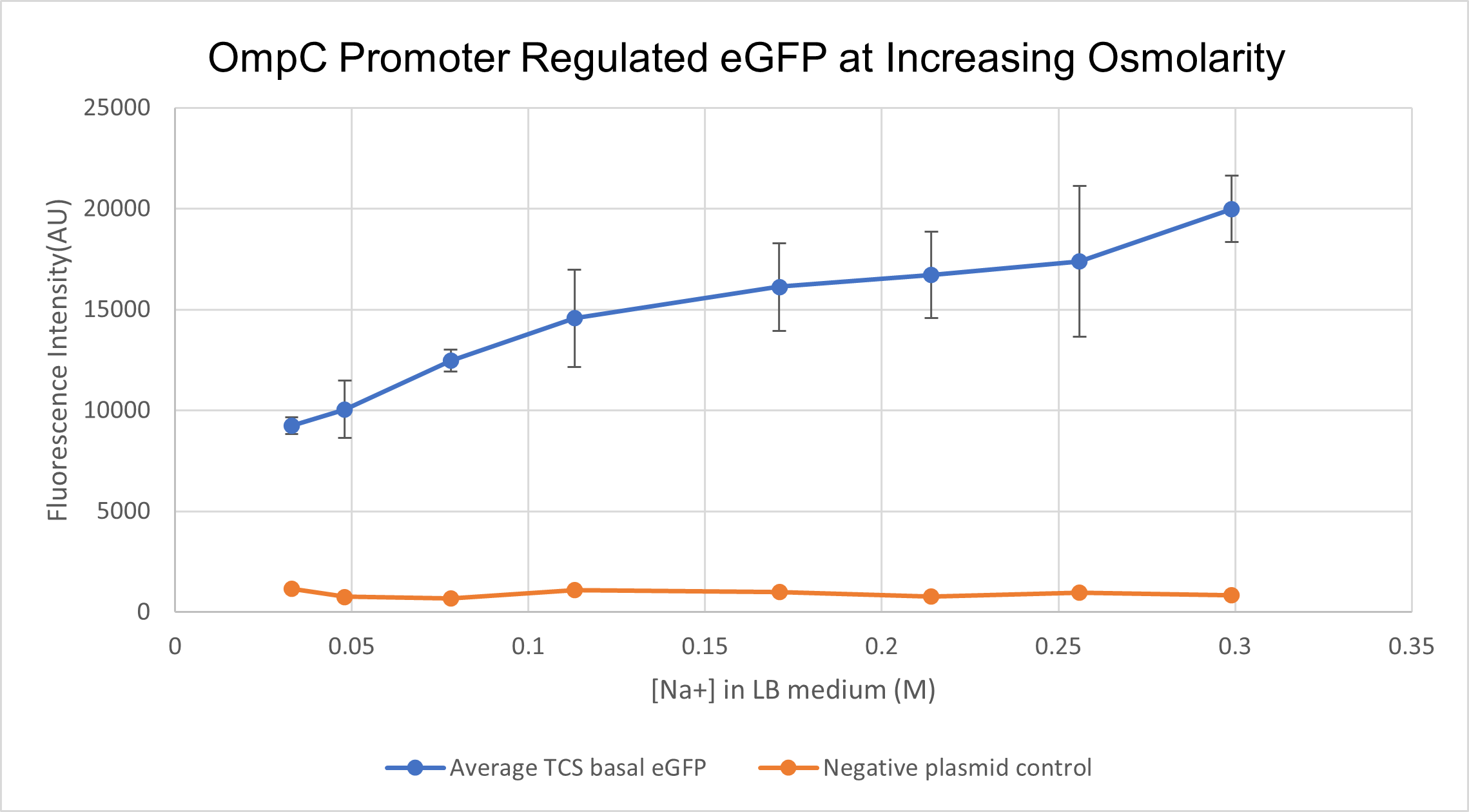
(Characterised by HKUST 2021)
Summary
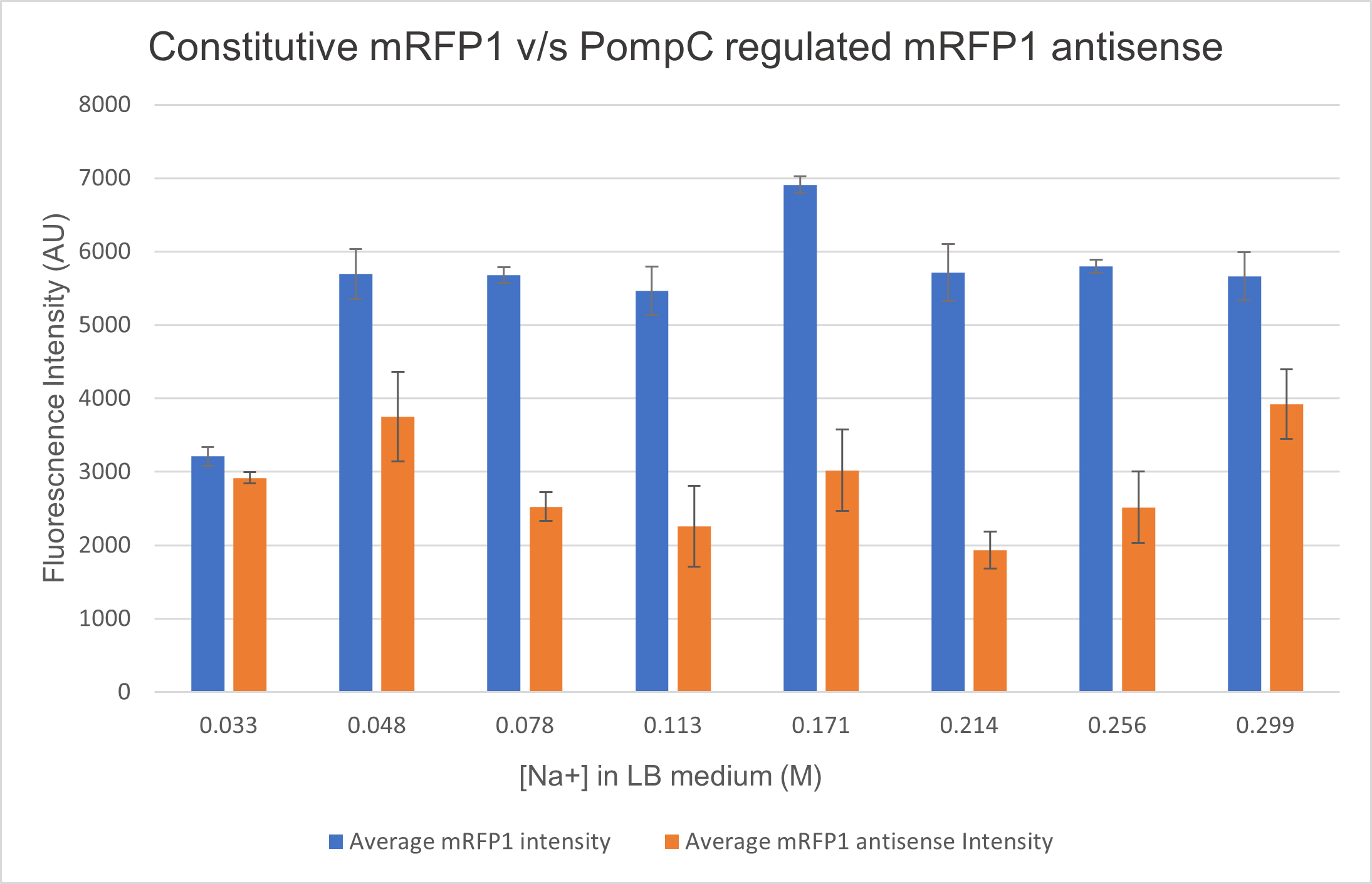
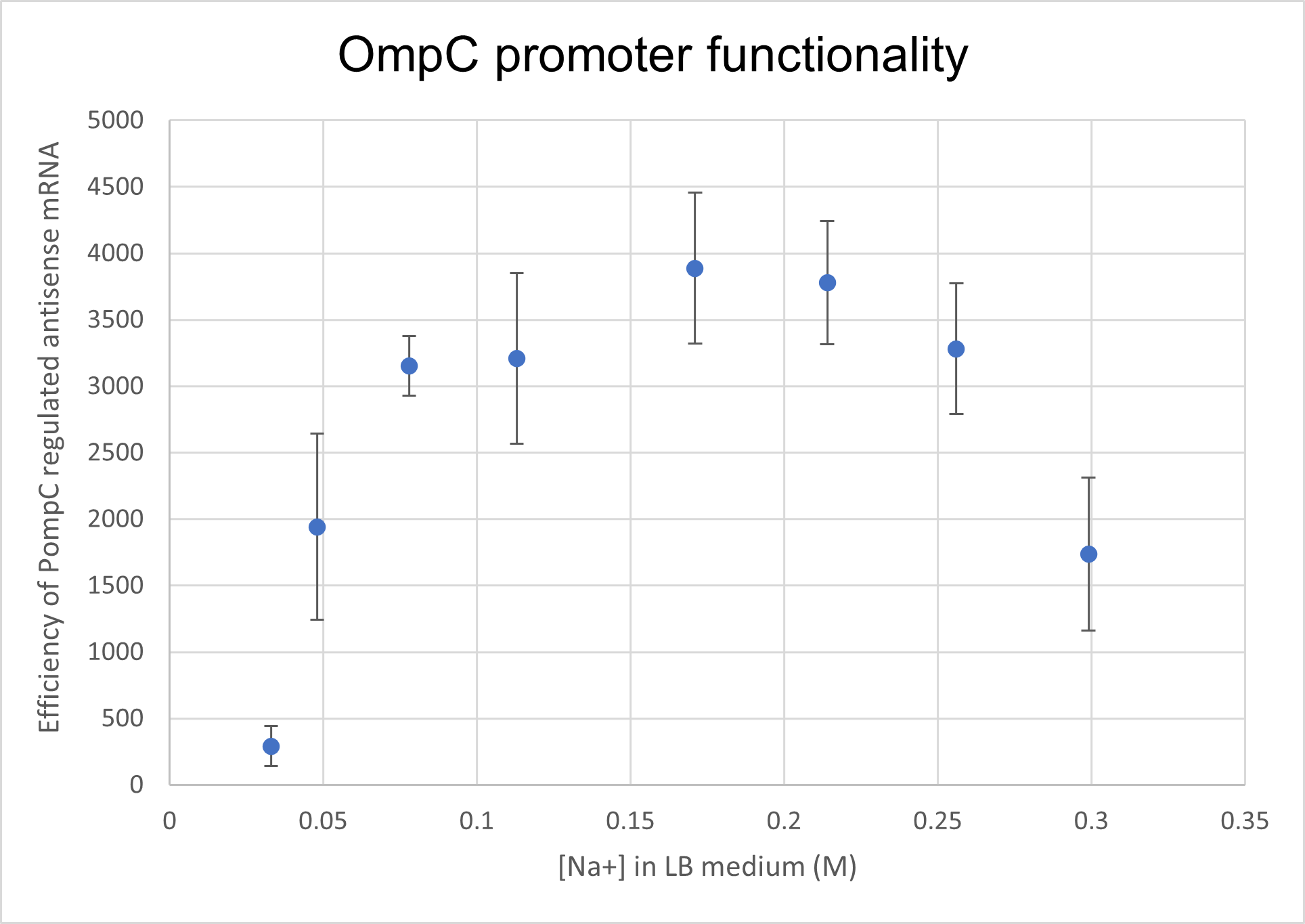
<p>We analysed the activity of OmpC promoter by regulating an antisense RNA against constitutively expressed mRFP1 (BBa_K4061095) and comparing the fluorescence intensity with that of only constitutively expressed mRFP1 with no antisense molecule. We computed the difference in the fluorescence intensities of both constructs and observed that the antisense RNA was effective at suppressing the constitutive fluorophore production at higher medium concentrations.Experiments
Two cells ( E. coli BL21) were transformed with 2 constructs- Constitutively expressed mRFP1 and Constitutively expressed mRFP1_pOmpC regulated antisense RNA. We cultured these cells in LB media of increasing NaCl concentration and observed the efficiency of the antisense RNA to suppress the fluorescence. The rationale was that at higher osmolarities, the OmpC promoter would be more active, producing more antisense RNA and hence suppressing more fluorescence from mRFP1.
Results and Discussion
As seen in the graph below, the PompC regulated antisense RNA indeed is able to suppress fluorescence at higher medium osmolarities . This supports the hypothesis since the antisense mRNA is under the regulation of the OmpC promoter, which is more active at higher osmolarities. However, surprisingly, the efficiency of the promoter or perhaps even the precision of measurement seemed to drop at higher osmolarities. It could either be that the cell was undergoing extreme stress at such high osmolarities and the death rate was higher, which renders the data beyond a 0.256M of NaCl in LB inconclusive.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
//classic/regulatory/uncategorized
//collections/probiotics/control
//direction/forward
//function/coliroid
//promoter
//regulation/positive
//rnap/prokaryote/ecoli/sigma70
| biology | |
| direction | Forward |
| negative_regulators | |
| positive_regulators | 1 |

 1 Registry Star
1 Registry Star