Difference between revisions of "Part:BBa K907000"
m |
|||
| Line 3: | Line 3: | ||
---- | ---- | ||
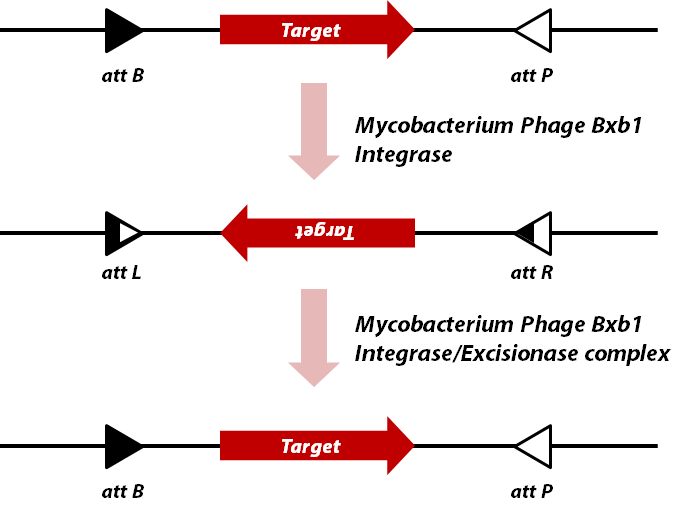
| − | <br> | + | <br> [[Image: KAIST_DNA_recombination_via_Bxb1_Int,_Xis.png |center|450px]] This part is a protein coding part which encodes |
| − | [[Image: KAIST_DNA_recombination_via_Bxb1_Int,_Xis.png |center|450px]] | + | Bxb1 gp35, which is a '''DNA integrase''' of ''Mycobacterium phage Bxb1''. |
| − | This part is a protein coding part which encodes Bxb1 gp35, which is a '''DNA integrase''' of ''Mycobacterium phage Bxb1''. | + | <br>The Bxb1 integrase is a DNA recombinase, more precisely a member of serine integrase family. It recognizes specific sequences, |
| − | <br>The Bxb1 integrase is a DNA recombinase, more precisely a member of serine integrase family. It recognizes specific sequences, called ''attB'' and ''attP'', and then integrates, inverts, or excises dsDNA depending on the orientation of recognition sequences. | + | called ''attB'' and ''attP'', and then integrates, inverts, or excises dsDNA depending on the orientation of recognition |
| − | We used this integrase to invert specific sequence in plasmid. The protein is well expressed in ''E.coli(strain MG1655)'' | + | sequences. We used this integrase to invert specific sequence in plasmid. The protein is well expressed in ''E.coli(strain |
| − | When it inverts DNA sequence, the ''attB'' and ''attP'' sequences are changed into'' attL'' and ''attR'', as other DNA recombinases do. Another protein called Bxb1 gp47([https://parts.igem.org/Part:BBa_K907002 BBa_K0907002]) binds to integrase-DNA complex and this complex flips inverted DNA back into original sequence by regenerating ''attB'' and ''attP'' sequences. | + | MG1655)'' When it inverts DNA sequence, the ''attB'' and ''attP'' sequences are changed into'' attL'' and ''attR'', as other |
| − | ---- | + | DNA recombinases do. Another protein called Bxb1 gp47([https://parts.igem.org/Part:BBa_K907002 BBa_K0907002]) binds to integrase-DNA |
| − | '''<Part Demonstration>''' | + | complex and this complex flips inverted DNA back into original sequence by regenerating ''attB'' and ''attP'' sequences. |
| − | <br>[[Image: KAIST Experimental Results.PNG |center|350px|thumb|'''Figure 1. Experimental results of BBa_K907000.''' ]] | + | ---- ''' |
| − | Two ep-tubes designated as BBa_K907004 are containing centrifuged E.coli MG1655 cells possessing BBa_K907004. pTrcHis2A vector containing <partinfo>BBa_K907000</partinfo>, controlled by Trc promoter, transformed into MG1655-BBa_K907004. The double transformed E.coli MG1655 cells showed color red rather than yellowish green color of MG1655-BBa_K907004. Two ep-tubes designated as BBa_K907005 are containing centrifuged E.coli MG1655 cells possessing BBa_K907005. pTrcHis2A vector containing <partinfo>BBa_K907000</partinfo>, controlled by Trc promoter, transformed into MG1655-BBa_K907005. The double transformed E.coli MG1655 cells showed pink color rather than intense red color of MG1655-BBa_K907005. Gathered cells are not showing perfect yellowish green color because initial state color of mRFP was too strong. | + | <Part Demonstration>''' |
| − | <br>Trc promoter, however, has basal level expression of Bxb1 gp35, Mycobacterium Phage Bxb1 DNA integrase. Even though the basal expression level is very low, the double transformants were available to change its color without IPTG induction. Thus we presume | + | <br>[[Image: KAIST Experimental Results.PNG |center|350px|thumb|'''Figure 1. Experimental results of BBa_K907000.''' ]] |
| − | <br>Further information are available at [http://2012.igem.org/Team:KAIST_Korea KAIST_iGEM_2012 Wiki] | + | Two ep-tubes designated as BBa_K907004 are containing centrifuged E.coli MG1655 cells possessing BBa_K907004. pTrcHis2A |
| − | ---- | + | vector containing |
| − | '''<Related Parts>''' | + | <partinfo>BBa_K907000</partinfo>, controlled by Trc promoter, transformed into MG1655-BBa_K907004. The double transformed E.coli MG1655 |
| − | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907001] - Mycobacterium Phage Bxb1 excisionase | + | cells showed color red rather than yellowish green color of MG1655-BBa_K907004. Two ep-tubes designated as BBa_K907005 are |
| − | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907002] - Binary Signal Generator, RBS(reverse) - attB - Promoter - attP - RBS | + | containing centrifuged E.coli MG1655 cells possessing BBa_K907005. pTrcHis2A vector containing |
| − | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907003] - Binary Signal Generator, Promoter Reversed, RBS(reverse) - attB - Promoter(reverse) - attP - RBS | + | <partinfo>BBa_K907000</partinfo>, controlled by Trc promoter, transformed into MG1655-BBa_K907005. The double transformed E.coli MG1655 |
| − | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907004] - Dual Phase Protein Generator(GFP default). mRFP and GFP | + | cells showed pink color rather than intense red color of MG1655-BBa_K907005. Gathered cells are not showing perfect yellowish |
| − | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907005] - Dual Phase Protein Generator(mRFP default). mRFP and GFP | + | green color because initial state color of mRFP was too strong. '''This results demonstrate that this part BBa_K907000 is |
| − | + | working as we expected.''' The cells were cultured in 100 mL LB media(1% Cm,AP each) and gathered incubating 12 hours more | |
| + | after induction with 1mM of IPTG. Culture condition was maintained at 37'C and 220 rpm. | ||
| + | <br>Trc promoter, however, has basal level expression of Bxb1 gp35, Mycobacterium Phage Bxb1 DNA integrase. Even though | ||
| + | the basal expression level is very low, the double transformants were available to change its color without IPTG induction. | ||
| + | Thus we presume that BBa_K907000 is very efficient in bacterial cells. To diminish the effect of basal transcription level, | ||
| + | we performed several optimizations. | ||
| + | <br>Further information are available at [http://2012.igem.org/Team:KAIST_Korea KAIST_iGEM_2012 Wiki] ---- ''' | ||
| + | <Related Parts>''' | ||
| + | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907001] - Mycobacterium Phage Bxb1 excisionase | ||
| + | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907002] - Binary Signal Generator, RBS(reverse) - attB - Promoter - attP | ||
| + | - RBS | ||
| + | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907003] - Binary Signal Generator, Promoter Reversed, RBS(reverse) - attB | ||
| + | - Promoter(reverse) - attP - RBS | ||
| + | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907004] - Dual Phase Protein Generator(GFP default). mRFP and GFP | ||
| + | <br>[https://parts.igem.org/Part:BBa_K907001 BBa_K907005] - Dual Phase Protein Generator(mRFP default). mRFP and GFP | ||
| − | <!-- Add more about the biology of this part here | + | <h1> Contribution</h1> |
| + | <ul> | ||
| + | <li>'''Group:''' ETH Zurich 2016 | ||
| + | <li>'''Author:''' Asli Azizoglu | ||
| + | <li>'''Summary:''' We cloned and characterised a codon optimised bxb1 for <i>E.coli</i>, and sent it to the registry as | ||
| + | a biobrick. Our biobrick can be found here with a fast degradation tag[[Part:BBa_K2116009]] and here without any degradation | ||
| + | tags[[Part:BBa_K2116026]]. | ||
| + | </ul> | ||
| + | |||
| + | <h1>Kinetic Characterisation</h1> | ||
| + | We investigated the kinetics of bxb1 flipping using flow cytometry. | ||
| + | <h2>Genetic Design</h2> | ||
| + | <ul> | ||
| + | <li> Bxb1 was expressed under a tet promoter, without any degradation tags. The RBS has a strenght of 1209.69 au (translation | ||
| + | rate), as calculated by the [https://salislab.net/software/ Salis RBS Calculator]. This construct was cloned on a medium | ||
| + | copy plasmid with the p15A replication of origin. | ||
| + | <li> TetR was expressed under the control of medium-strength constitutive promoter [[Part:Bba_J23118]], and cloned onto a | ||
| + | low copy plasmid with pSC101/Rep101 replication of origin. | ||
| + | <li> We also constructed a dual fluorescence reporter system [[Part:BBa_K2116024]] that expresses GFP when bxb1 flips a directional | ||
| + | promoter. | ||
| + | </ul> | ||
| + | <h2 id="experimental_setup"> Experimental Setup </h2> | ||
| + | <ul> | ||
| + | <li> The test construct with bxb1 was transformed together with the reporter construct, and either with TetR or with the empty | ||
| + | backbone as <b>control</b>. | ||
| + | <li> Cells were grown in LB medium for 4 hours, and then transfered to minimal M9 medium with a 1:100 dilution. aTc was added | ||
| + | at given concentrations at OD<sub>600</sub>. Samples were taken at the specified time points, spun down and resuspended | ||
| + | in PBS at OD<sub>600</sub> 0.01 and kept on ice until measurement. | ||
| + | <li> Results are presented as mean fluorescence per cell and the standard error of the mean. | ||
| + | </ul> | ||
| + | |||
| + | <h2>Results</h2> | ||
| + | <p>We observed that maximum flipping is reached at around 8h under these conditions. The response correlates with aTc concentration. | ||
| + | However compared to the control, where there is no TetR, we see 6-fold less flipping. This could be improved by increasing | ||
| + | the concentration of aTc, however we have found that at 8000 ng/μL cell growth was inhibited.</p> | ||
| + | |||
| + | [[File:T--ETH Zurich--bxb1kineticsfull.png|500px|thumb|center|Time and dose response of bxb1 flipping a directional promoter. | ||
| + | Induction at OD<sub>600</sub> 0.5 with aTc at given concentrations. Data measured by flow cytometry at given time points. | ||
| + | Values are shown as mean fluorescence per cell, error bars indicate SEM. ]] | ||
| + | |||
| + | <h1> Comparison between Codon Optimised and Registry Bxb1 Integrase </h1> | ||
| + | <p>We compared the kinetics of our codon optimised bxb1 [[Part:BBa_K2116026]] and the one found previously on the registry | ||
| + | [[Part:BBa_K907000]] that was not codon optimised. The same genetic and experimental setup was used as above. The only | ||
| + | difference between the two constructs is that the non-codon optimised bxb1 has a weaker RBS, with a strength of 251.82au | ||
| + | (translation rate), as calculated by the [[https://salislab.net/software Salis RBS Calculator]]. Due to this difference | ||
| + | in construction we compared each bxb1 to it's own control (see above, [[#experimental_setup|Experimental Setup]]).</p> | ||
| + | <p> We could show that the codon optimised bxb1 could reach ~1/6th flipping compared to control, whereas the non-codon optimised | ||
| + | one reached only ~1/260th of its own control. We thus argue that the codon optimised bxb1 is more efficient, especially | ||
| + | since the fold increase in efficiency is a lot more than the fold increase in RBS strenght. </p> | ||
| + | |||
| + | [[File:T--ETH Zurich--bxb1kineticsfull.png|500px|thumb|center|Comparison of codon optimised and non-optimised bxb1 flipping | ||
| + | efficiency. Test constructs have bxb1 expressed under the Tet promoter, with TetR present in the system. Control doesn't | ||
| + | have TetR in the system, and thus constitutively expresses bxb1. While non-codon optimised bxb1 can only reach ~1/260th | ||
| + | of the full flipping reached by control, the codon optimised one reaches ~1/6th of its own control. Induction at OD<sub>600</sub> 0.5 with 2000ng/μL. Data measured by flow cytometry at 16h. Values are shown as mean fluorescence per cell, error bars | ||
| + | indicate SEM. ]] | ||
| + | |||
| + | |||
| + | |||
| + | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
<!-- --> | <!-- --> | ||
| − | <span class='h3bb'>Sequence and Features</span> | + | <span class='h3bb'>Sequence and Features</span> |
| − | <partinfo>BBa_K907000 SequenceAndFeatures</partinfo> | + | <partinfo>BBa_K907000 SequenceAndFeatures</partinfo> |
| − | <!-- Uncomment this to enable Functional Parameter display | + | <!-- Uncomment this to enable Functional Parameter display |
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K907000 parameters</partinfo> | <partinfo>BBa_K907000 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
Revision as of 18:10, 22 October 2016
Mycobacterium Phage Bxb1 gp35, DNA integrase
This part is a protein coding part which encodes
Bxb1 gp35, which is a DNA integrase of Mycobacterium phage Bxb1.
The Bxb1 integrase is a DNA recombinase, more precisely a member of serine integrase family. It recognizes specific sequences,
called attB and attP, and then integrates, inverts, or excises dsDNA depending on the orientation of recognition
sequences. We used this integrase to invert specific sequence in plasmid. The protein is well expressed in E.coli(strain
MG1655) When it inverts DNA sequence, the attB and attP sequences are changed into attL and attR, as other
DNA recombinases do. Another protein called Bxb1 gp47(BBa_K0907002) binds to integrase-DNA
complex and this complex flips inverted DNA back into original sequence by regenerating attB and attP sequences.
<Part Demonstration>
Two ep-tubes designated as BBa_K907004 are containing centrifuged E.coli MG1655 cells possessing BBa_K907004. pTrcHis2A
vector containing
BBa_K907000, controlled by Trc promoter, transformed into MG1655-BBa_K907004. The double transformed E.coli MG1655
cells showed color red rather than yellowish green color of MG1655-BBa_K907004. Two ep-tubes designated as BBa_K907005 are
containing centrifuged E.coli MG1655 cells possessing BBa_K907005. pTrcHis2A vector containing
BBa_K907000, controlled by Trc promoter, transformed into MG1655-BBa_K907005. The double transformed E.coli MG1655
cells showed pink color rather than intense red color of MG1655-BBa_K907005. Gathered cells are not showing perfect yellowish
green color because initial state color of mRFP was too strong. This results demonstrate that this part BBa_K907000 is
working as we expected. The cells were cultured in 100 mL LB media(1% Cm,AP each) and gathered incubating 12 hours more
after induction with 1mM of IPTG. Culture condition was maintained at 37'C and 220 rpm.
Trc promoter, however, has basal level expression of Bxb1 gp35, Mycobacterium Phage Bxb1 DNA integrase. Even though
the basal expression level is very low, the double transformants were available to change its color without IPTG induction.
Thus we presume that BBa_K907000 is very efficient in bacterial cells. To diminish the effect of basal transcription level,
we performed several optimizations.
Further information are available at [http://2012.igem.org/Team:KAIST_Korea KAIST_iGEM_2012 Wiki] ----
<Related Parts>
BBa_K907001 - Mycobacterium Phage Bxb1 excisionase
BBa_K907002 - Binary Signal Generator, RBS(reverse) - attB - Promoter - attP
- RBS
BBa_K907003 - Binary Signal Generator, Promoter Reversed, RBS(reverse) - attB
- Promoter(reverse) - attP - RBS
BBa_K907004 - Dual Phase Protein Generator(GFP default). mRFP and GFP
BBa_K907005 - Dual Phase Protein Generator(mRFP default). mRFP and GFP
Contribution
- Group: ETH Zurich 2016
- Author: Asli Azizoglu
- Summary: We cloned and characterised a codon optimised bxb1 for E.coli, and sent it to the registry as a biobrick. Our biobrick can be found here with a fast degradation tagPart:BBa_K2116009 and here without any degradation tagsPart:BBa_K2116026.
Kinetic Characterisation
We investigated the kinetics of bxb1 flipping using flow cytometry.
Genetic Design
- Bxb1 was expressed under a tet promoter, without any degradation tags. The RBS has a strenght of 1209.69 au (translation rate), as calculated by the Salis RBS Calculator. This construct was cloned on a medium copy plasmid with the p15A replication of origin.
- TetR was expressed under the control of medium-strength constitutive promoter Part:Bba_J23118, and cloned onto a low copy plasmid with pSC101/Rep101 replication of origin.
- We also constructed a dual fluorescence reporter system Part:BBa_K2116024 that expresses GFP when bxb1 flips a directional promoter.
Experimental Setup
- The test construct with bxb1 was transformed together with the reporter construct, and either with TetR or with the empty backbone as control.
- Cells were grown in LB medium for 4 hours, and then transfered to minimal M9 medium with a 1:100 dilution. aTc was added at given concentrations at OD600. Samples were taken at the specified time points, spun down and resuspended in PBS at OD600 0.01 and kept on ice until measurement.
- Results are presented as mean fluorescence per cell and the standard error of the mean.
Results
We observed that maximum flipping is reached at around 8h under these conditions. The response correlates with aTc concentration. However compared to the control, where there is no TetR, we see 6-fold less flipping. This could be improved by increasing the concentration of aTc, however we have found that at 8000 ng/μL cell growth was inhibited.
Comparison between Codon Optimised and Registry Bxb1 Integrase
We compared the kinetics of our codon optimised bxb1 Part:BBa_K2116026 and the one found previously on the registry Part:BBa_K907000 that was not codon optimised. The same genetic and experimental setup was used as above. The only difference between the two constructs is that the non-codon optimised bxb1 has a weaker RBS, with a strength of 251.82au (translation rate), as calculated by the [Salis RBS Calculator]. Due to this difference in construction we compared each bxb1 to it's own control (see above, Experimental Setup).
We could show that the codon optimised bxb1 could reach ~1/6th flipping compared to control, whereas the non-codon optimised one reached only ~1/260th of its own control. We thus argue that the codon optimised bxb1 is more efficient, especially since the fold increase in efficiency is a lot more than the fold increase in RBS strenght.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 192
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 466
Illegal XhoI site found at 553 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1105
Illegal NgoMIV site found at 1192
Illegal AgeI site found at 242 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1300