Difference between revisions of "Part:BBa K592010"
(→Usage and Biology) |
(→Usage and Biology) |
||
| Line 46: | Line 46: | ||
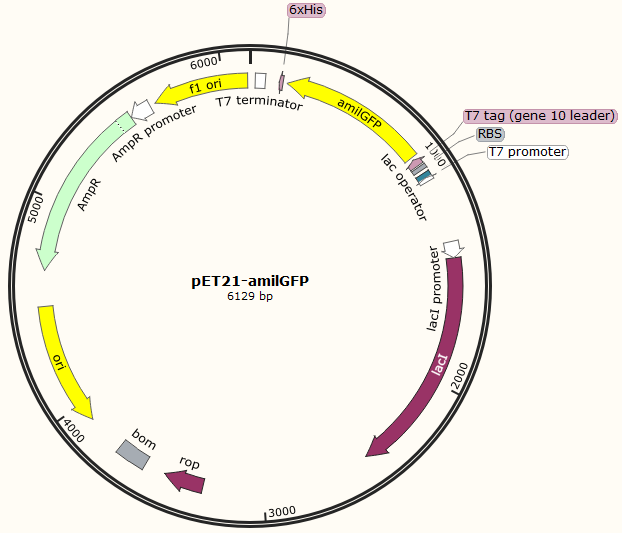
This part was characterized in the measurement of amilGFP's absorption and emission spectra by the iGEM2019 SCU-China team. To express this chromoprotein, we cloned this part downstream T7 promoter on pET21α vector and transformed this T7-amilGFP-pET21α into E.coli BL21 cells.(Figure 1) | This part was characterized in the measurement of amilGFP's absorption and emission spectra by the iGEM2019 SCU-China team. To express this chromoprotein, we cloned this part downstream T7 promoter on pET21α vector and transformed this T7-amilGFP-pET21α into E.coli BL21 cells.(Figure 1) | ||
| + | [[File:measurement 1.png|300px|]] | ||
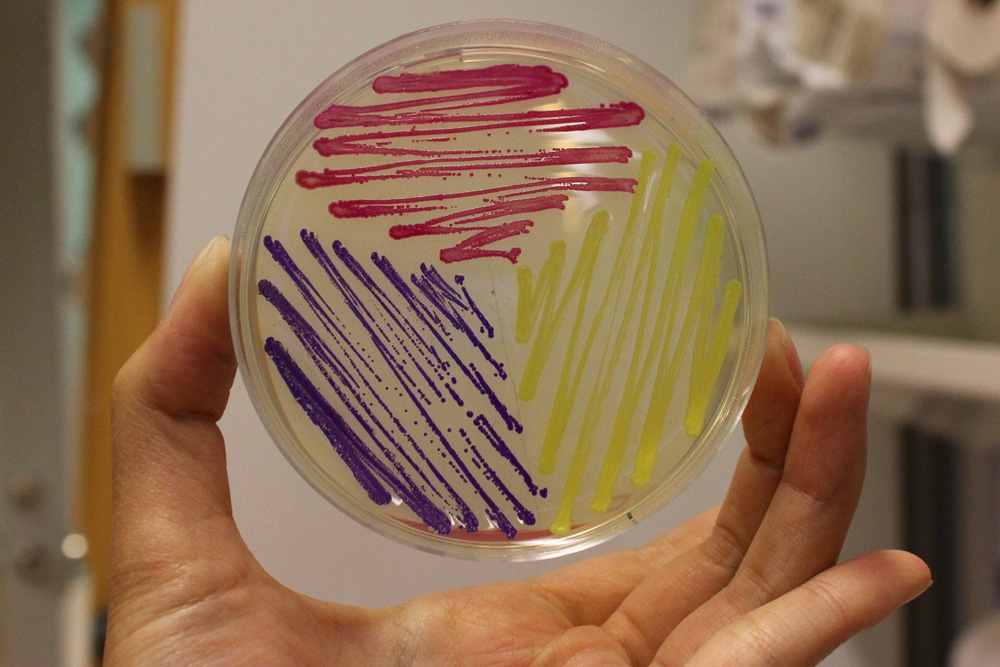
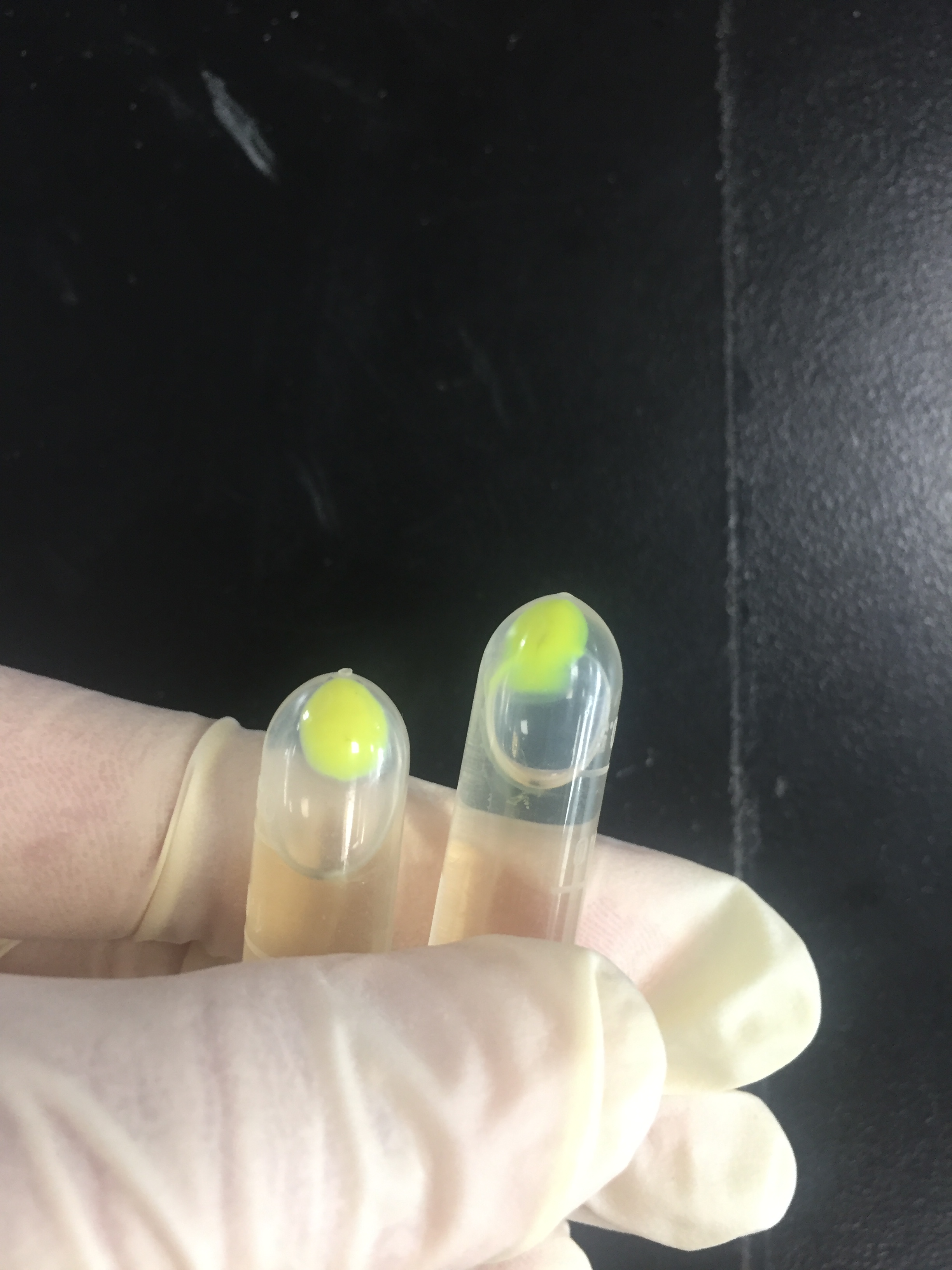
The amilGFP protein was visible in yellow colour less than 24-hour incubation after adding IPTG both on LB plates or in liquid culture.(Figure 2) | The amilGFP protein was visible in yellow colour less than 24-hour incubation after adding IPTG both on LB plates or in liquid culture.(Figure 2) | ||
Revision as of 07:16, 15 October 2019
amilGFP, yellow chromoprotein
This chromoprotein from the coral Acropora millepora, amilGFP, naturally exhibits strong yellow color when expressed. The color is readily visible to naked eye both in LB-culture and on agar plates. Color development can be seen in less than 24 hours of incubation.
Important: This part is not available in the registry yet, however, the same part is available from the registry with the standard RBS B0034: BBa_K1033931.
Usage and Biology
This part is useful as a reporter.
Peking iGEM 2016 has fused this part with triple spytag. The fused protein is participate in Peking’s polymer network. By adding this protein, the whole polymer network become visible in most conditions. If you want to learn more about Peking’s polymer network and the role of mRFP in this network, please click here https://parts.igem.org/Part:BBa_K1989003", https://parts.igem.org/Part:BBa_K1989045" or https://parts.igem.org/Part:BBa_K1989048".
iGEM11_Uppsala-Sweden: Expression of chromoproteins. The images above show E coli constitutively expressing amilGFP BBa_K592010 (yellow), amilCP BBa_K592009 (blue), and RFP BBa_E1010 (red).
iGEM12_Uppsala_University: The Uppsala chromoprotein collection and RFP. The image shows pellets of E coli expressing chromoproteins eforRed BBa_K592012, RFP BBa_E1010, cjBlue BBa_K592011, aeBlue BBa_K864401, amilGFP BBa_K592010 and amilCP BBa_K592009.
iGEM17_SJTU_BioX_Shanghai:
J23119+Target3+amilGFP BBa_K2285013
J23119+Target1+amilGFP BBa_K2285017
An improved part has been constructed. Since this part is a coding sequence, we added a RBS which is on the upstream of sfGFP BBa_K515005 and terminator BBa_B1006 by PCR. What's more, our device have a stem-loop structure(called Target) following the constitutive promoter J23119, to achieve further control of amilGFP expression.
iGEM18_Bulgaria:
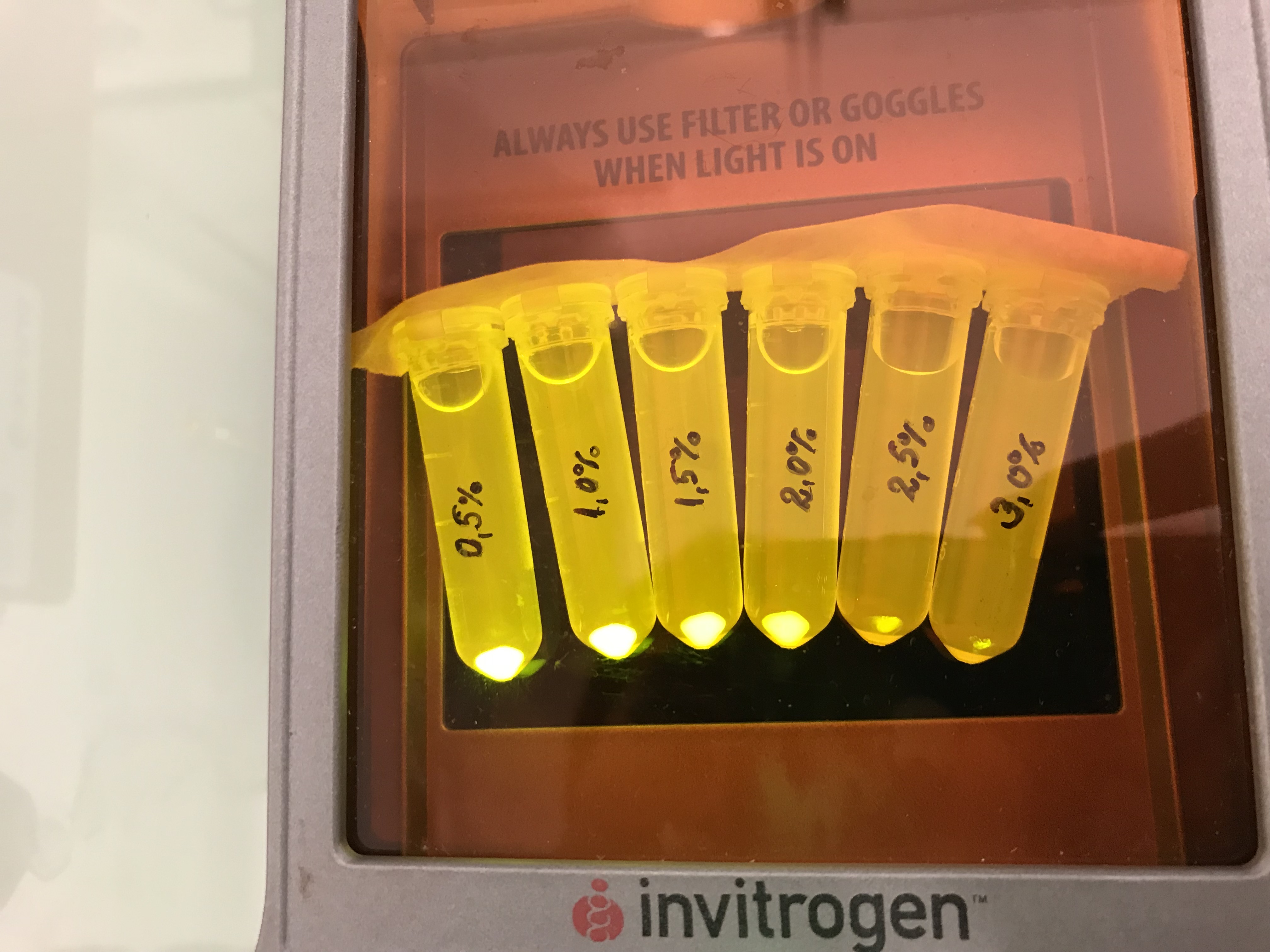
This part was characterized by the iGEM Bulgaria 2018 team. We cloned this chromoprotein CDS into a high copy number vector pSB1C3 that contains part BBa_K608002 (strong promoter and strong RBS). Additionally similar constructs with amilCP, cjBlue, gfasPurple, eforRed and spisPink were prepared. We tested all these constructs for colour stability upon re-cultivation. Under these conditions, most of the proteins showed as a severe metabolic burden to the host cells, leading to small sizes of the colonies and color loss upon re-cultivation. The exception was the amilGFP protein - it was stable enough to be used under the control of a strong promoter and on a high copy number vector. We also measured its growth kinetics in comparison with the standard pSB1C3 vector (with a red colour device) and a pSB1C3 vector with an AmilGFP CDS (without promoter and RBS, used as a control). The obtained data were normalized to AmilGFP-CDS vector (value of 1.00). The growth rate of pSB1C3-Amil GFP is 0.52 and the pSB1C3-mRFP red colour device (not with a strong promoter) has a value of 0,85. Despite these results we found that the AmilGFP was the only chromoprotein from the group tested that was always stable upon re-cultivation.
Next we tested the AmilGFP expression from our construct in LB media with increased levels of NaCl. The results can be seen on the following figure:
iGEM19_SCU-China:
This part was characterized in the measurement of amilGFP's absorption and emission spectra by the iGEM2019 SCU-China team. To express this chromoprotein, we cloned this part downstream T7 promoter on pET21α vector and transformed this T7-amilGFP-pET21α into E.coli BL21 cells.(Figure 1)
The amilGFP protein was visible in yellow colour less than 24-hour incubation after adding IPTG both on LB plates or in liquid culture.(Figure 2)
In order to measure the absorption and emission spectra of amilGFP, we cultured engineered E.coli cells in liquid LB medium, induced the expression of amilGFP with IPTG and incubated at 37℃ overnight. After centrifugation, Cells were treated with 5ml PBS, disrupted through ultrasonication and then centrifuged again, the supernatant was transferred into a new tube and measured with the Microplate Reader in a range between 300-700 nm. The absorption peak is 502 nm and the emission peak was at 470 nm.(Figure 3)
References
[http://www.ncbi.nlm.nih.gov/pubmed/18648549] Alieva, N. O., et al. 2008. Diversity and evolution of coral fluorescent proteins. PLoS One 3:e2680.
GenBank: AY646067.1
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]