Part:BBa_K3823012
hydrogen sulfide sensitive, three-gear adjustable kill switch
Design
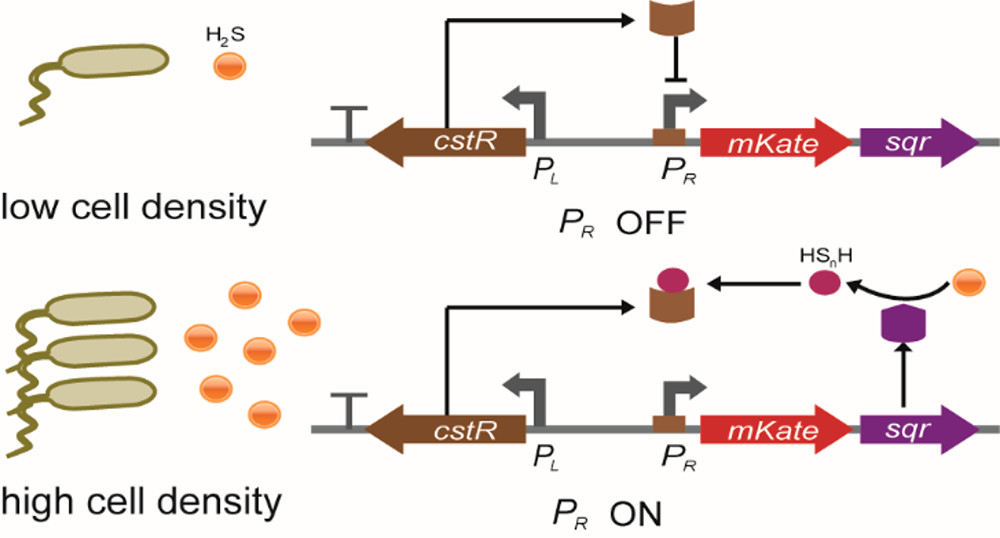
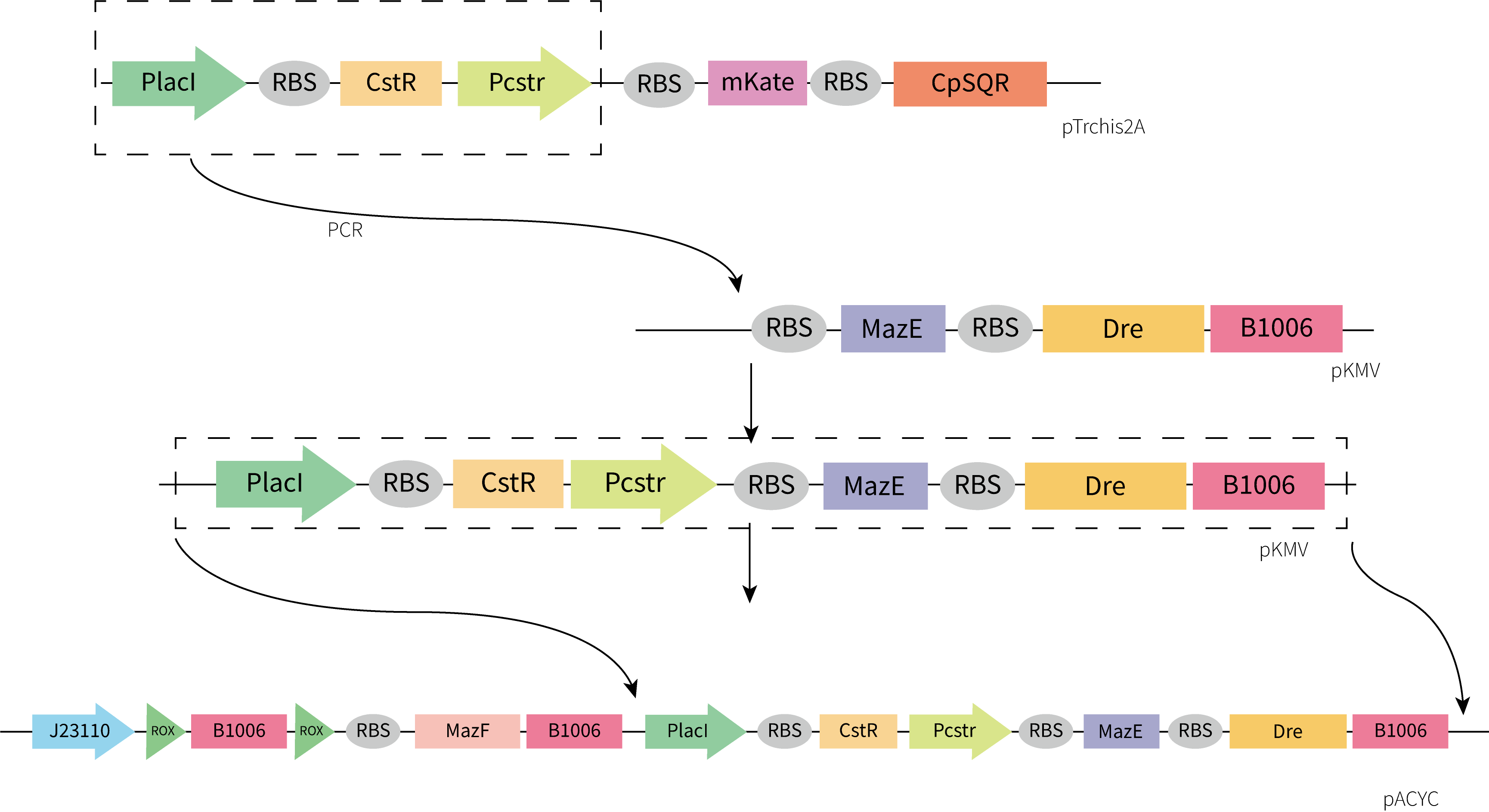
Biosafety has always been an important part in iGEM. To prevent our engineered bacteria from leaking into the environment, we designed a H2S sensitive three-gear kill switch to ensure this. H2S is an important factor in the working environment of our engineered bacteria, so we chose to use H2S concentration to control the on-off of the kill switch. By searching for the biosensors of various gas molecules, we found a HSnH-sensing biosensor (the product of SQR oxidized H2S happens to be HSnH) -- a gene regulator, CstR, which can sense HSnH and turn on the expression of the downstream genes (Figure 1.)[1].
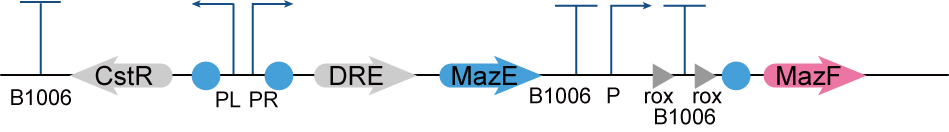
If we design a two-gear switch, namely, the switch is off when H2S concentration is high and on when H2S concentration is low, then we need to maintain high concentration of H2S in the culture environment, but this is obviously not realistic. Therefore, we designed a three-gear kill switch based on the Dre/rox recombinant enzyme system [2](referring to the work of the [http://2017.igem.org/Team:Edinburgh_UG Edinburgh UG in 2017] and the MazEF toxin-antitoxin system[3], which can achieve more flexible regulation of the survival of our engineered bacteria. (Figure 2.)
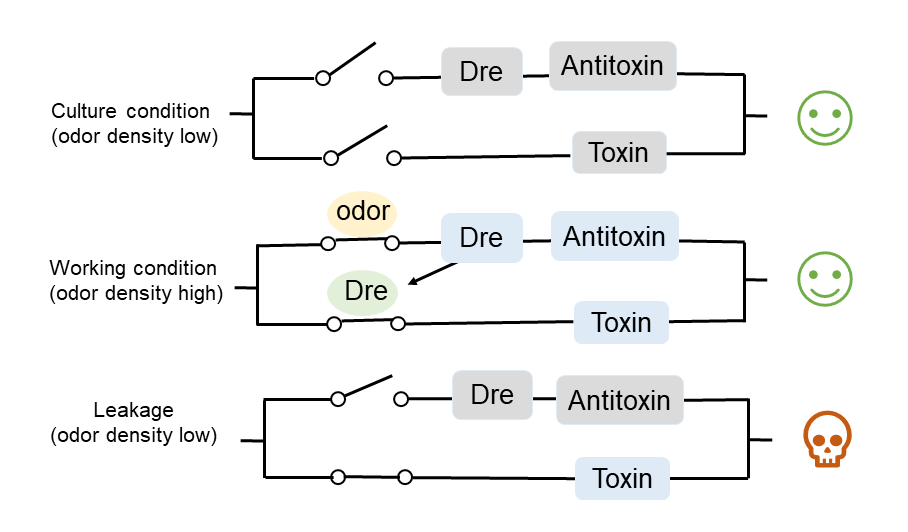
Under the culture condition, H2S concentration is not high enough to turn on any of the three pathways, so the bacteria will grow normally. When the concentration of H2S is high, H2S will remove the inhibition of CstR on the downstream gene expression by combining with it, then Dre will be expressed, and the terminator before MazF will be removed, resulting in the expression of MazF toxin. Meanwhile, due to the expression of the antitoxin MazE, the bacteria will not die. When working bacteria leak into the environment, H2S concentration in the environment will return to the normal (low) level, the inhibition of CstR will not be removed, and MazE will no longer be expressed. At this time, because MazF has been expressed, a large amount of MazF will kill the bacteria over time. (Figure 3.)
Experiments
Plasmid construction
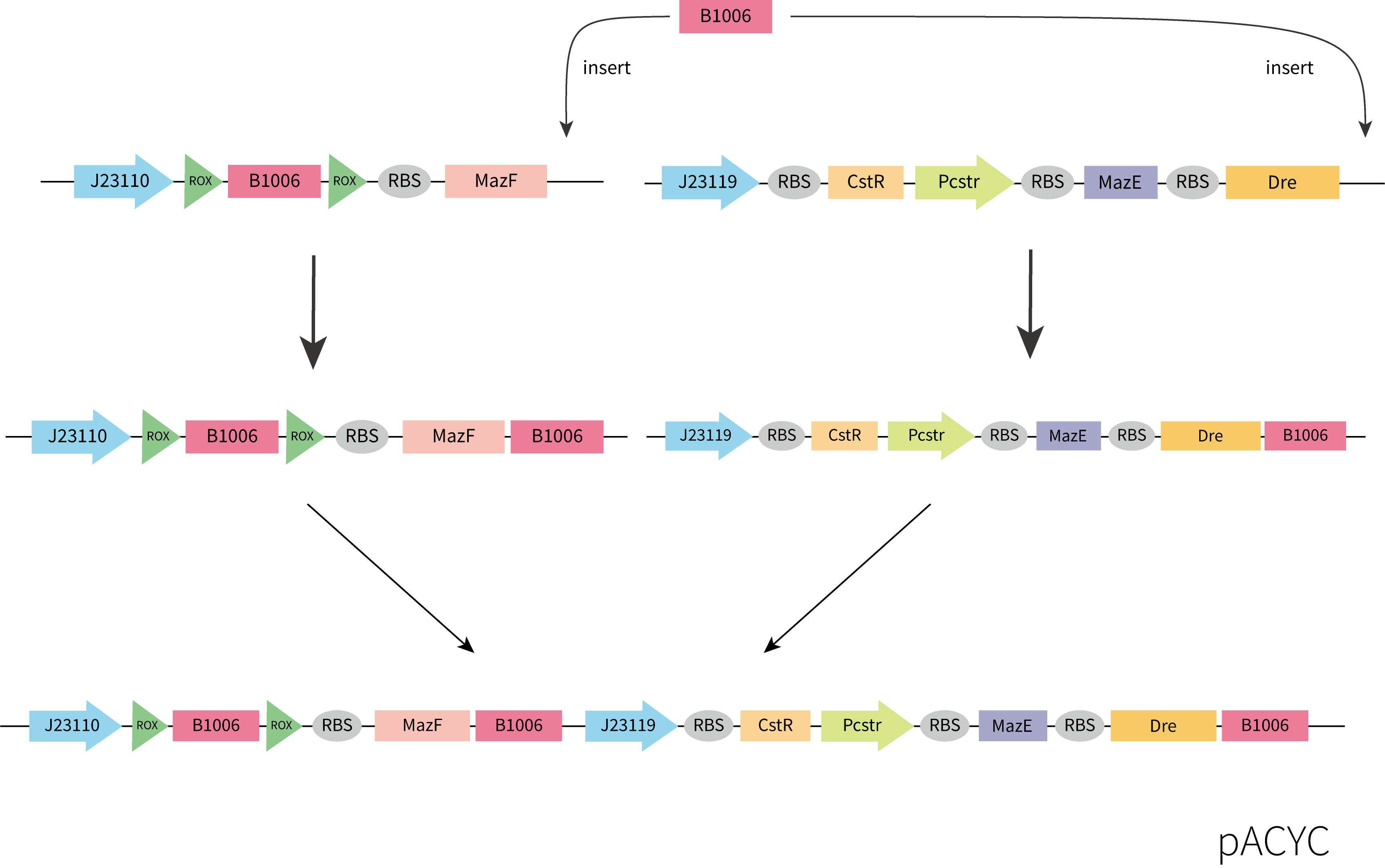
Considering the accessibility of the template and the time limitation, we adopted the strategy of assembling the functional components separately obtained by gene synthesis. Considering plasmid incompatibility, we chose pACYC as the vector for this part, which has a different replication system from pET-3a. Plasmids were constructed by restriction enzyme digestion and ligation and In-Fusion. The construction process we designed is shown below.
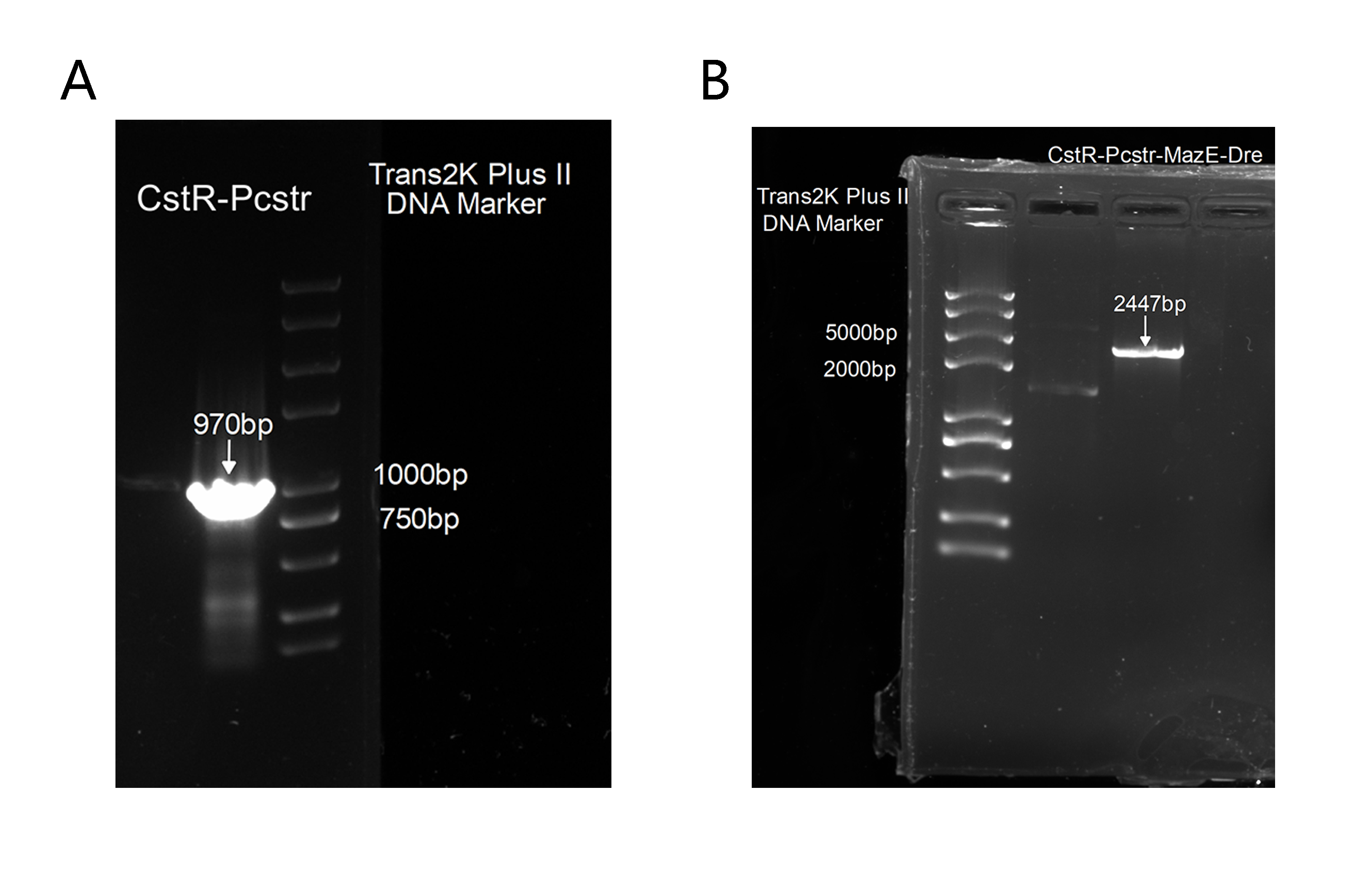
Unfortunately, the synthesis of J23119-CstR-Pcstr-MazE -Dre encountered difficulty so that we couldn't obtain the sequence of J23119-CstR-Pcstr. Considering this part was very necessary for our work, we contacted Professor Huaiwei Liu from Shandong University based on literature and fortunately got his help. Using the plasmid he donated as a template, we successfully amplified the CstR-Pcstr sequence(Figure 6A) and used it for our plasmid construction.
We successfully constructed the plasmid CstR-Pcstr-MazE -Dre and amplified CstR-Pcstr-MazE-Dre(Figure 6B).Then we cloned CstR-Pcstr-MazE-Dre to pACYC-rox-ter-rox-MazF.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 1300
Illegal PstI site found at 919
Illegal PstI site found at 1607 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 1300
Illegal NheI site found at 7
Illegal NheI site found at 30
Illegal PstI site found at 919
Illegal PstI site found at 1607 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 1300
Illegal BglII site found at 896
Illegal BglII site found at 2482
Illegal BamHI site found at 36
Illegal BamHI site found at 543
Illegal BamHI site found at 1660
Illegal BamHI site found at 2996
Illegal XhoI site found at 2954 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 1300
Illegal PstI site found at 919
Illegal PstI site found at 1607 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 1300
Illegal PstI site found at 919
Illegal PstI site found at 1607 - 1000COMPATIBLE WITH RFC[1000]
| None |