Part:BBa_K3755041
CMV enhancer+CMV promoter+GCaMP6m+SV40 PolyA signal
This composite part is constructed to express GCaMP6m in mammalian cells.
Usage and Biology
This part is cloned in pEGFP, that's why CMV enhancer, CMV promoter and SV40 PolyA signal are used. GCaMP6m is expressed in this composite part. GCaMP6m is a genetically encoded calcium indicator. When intracellular calcium concentration increases, emitted light at about 510nm will increase obviously. To see more detailed information about GCaMP6m, please go to view Part:BBa_K3755007. In our part, we used GCaMP6m to test whether calciums flow in cells after mechanical stimulation as we expected.
Experiment and result
Molecular cloning
We got [http://www.addgene.org/100838/ pAAV.Syn.Flex.GCaMP6m.WPRE.SV40](Part:BBa_K3755032)which contains GCaMP6m from Wei Shen'lab, Shanghaitech university. We used PCR to obtain DNA fragments of GCaMP6m. Then we inserted it behind CMV promoter using pEGFP as the backbone. Gibson assembly was used during the construction of this plasmid. The map of this plasmid is shown below.
Cotransfection and Stimulation
We cotransfected Piezo1.1(Part:BBa_K3755006) and GCaMP6m(Part:BBa_K3755007) into HEK293 cells. GCaMP6m is a calcium indicator. If calciums flow into the cotransfected cells through Piezo1.1, a raise of green fluorescence will be observed. The following figures show that we cotransfected these two plasmids successfully.
Protocol of cotransfection
1.Dilute 12.5 μl Lipofectamine 2000 Reagent in 125 μl Opti-MEM Medium and mix thoroughly.
2.Dilute 1.25 μg plasmid1 and 1.25 μg plasmid2 in 125 μl Opti-MEM Medium and mix thoroughly.
3.Add diluted plasmid to diluted Lipofectamine 2000 Reagent (1:1 ratio).
4.Incubate for at least 5 minutes at room temperature.
5.Digest all the 293 cells which are 90% confluent in a 10 cm (diameter) cell culture dish and suspend cells with 1 ml DMEM (10%FBS).
6.Take out 200 μl cell suspension and dilute it into 5 ml with DMEM (10%FBS).
7.Slowly drop the plasmid-lipid complex into the 5 ml cell suspension and softly mix thoroughly.
8.Plate the cell suspension into 24 well cell culture dish--1 ml each well.
9.Incubate cells for 48h at 37℃.
10.Analyze transfection efficiency in use of fluorescence microscope.
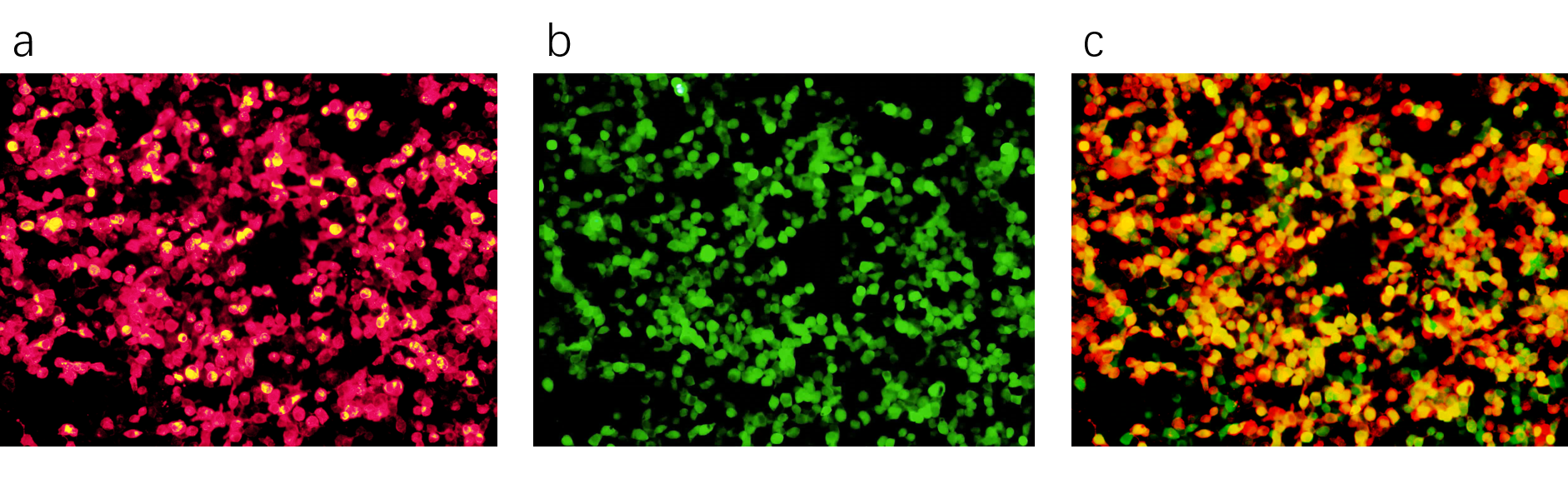
We used two different ways to stimulate Piezo1.1, and a control group is also built. The related results are shown below.
1.Yoda1
1ml 100uM Yoda1 was added to 1ml culture medium, so the final consentration of Yoda1 is 50uM. This proccess was finished under the inverted fluorescence microscope. A significant fluorescence change was observed in seconds.
Video1:Fluorescence change of cells under the stimulation of 50uM Yoda1, exposure time: 90ms, 20X objective
File:Yoda 8times.mp4
2.Shear stress
We gave the HEK293 cells the shear stress by sucking out their culture medium with a pipette. The change of fluorescence is faster and more obvious than that caused by Yoda1.
Video2:Faster fluorescence change of cells under the stimulation of shear stress, exposure time: 90ms, 20X objective
File:Force 4times.mp4
3.Control
In the control group, only GCaMP6m is transfected into the HEK293 cells. We also used 50uM of Yoda and shear stress to stimulate the cells in the control group. But no obvious change of fluorescence is observed in the control group under the condition of Yoda1 or shear stress.
Video3:Fluorescence change of cells in control group, exposure time: 90ms, 20X objective
File:Control 1.mp4
We also did a quantitative analysis of fluorescence changes. A program based on MATLAB was used to finish this quantitative analysis.
The curve showed the characteristics of rising first and then falling under the condition of Yoda1.(Figure3a) This is important to show a decreased fluorescence intensity, which is consistent with the features of Piezo1.1. The control group showed no obvious fluorescence change.(Figure3b)
These results show that GCaMP6m indicates the changes of intracellular calcium concentration successfully.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 542
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 683
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1524
Illegal SapI.rc site found at 1430
| None |


