Part:BBa_K3755006
mouse Piezo1.1
A splicing variant of mPiezo1 which is more sensitive to mechanical stimulation.
Usage and Biology
Introduction of Piezo1
Piezo1.1 is a splicing variant of Piezo1(BBa_K3281005). Piezo1 is a eukaryotic cation-selective mechanosensitive ion channel. While Piezo1 is stimulated by mechanical force, cations, especially calcium will flow into the cytoplasm. It will cause a significant increase in the concentration of calcium in the cytoplasm. Piezo1 plays an important role in many key biological activities, including blood pressure regulation, bone formation, and breathing. Piezo1 can also be activated by some small molecule compounds, like Yoda 1.
Difference between Piezo1.1 and Piezo1
Piezo1.1 lacks the alternatively spliced exon 30 of Piezo1. Exon 30 contains 72 nucleotides and encodes 24 amino acids from 1382 to 1405 of the mouse Piezo1. Compared with Piezo1, Piezo1.1 lacks both the lateral and central plug domains, which cause an increase of single-channel conductance and a decrease of Ca^2+^ permeability. So Piezo1.1 can be regarded as a more mechanosensitive mutant of Piezo1. We selected Piezo1.1 to be the mechanical sensor in our project.
Usage
This part can be used in a mechanosensitive gene expression system. Our team built up a part collection to achieve that. You can view this page for further information.
Experiment and result
Transfection
Piezo1.1 and mRuby2 are fused, so the red fluorescence in the field represents the expressed Piezo1.1. The following picture shows that we transfected Piezo1.1 into HEK293 cells successfully.
Cotransfection and Stimulation
We cotransfected Piezo1.1 and GCaMP6m(Part:BBa_K3755007) into HEK293 cells. GCaMP6m is a calcium indicator. If calciums flow into the cotransfected cells through Piezo1.1, a raise of green fluorescence will be observed. The following figures show that we cotransfected these two plasmids successfully.
Protocol of cotransfection
1.Dilute 12.5 μl Lipofectamine 2000 Reagent in 125 μl Opti-MEM Medium and mix thoroughly.
2.Dilute 1.25 μg plasmid1 and 1.25 μg plasmid2 in 125 μl Opti-MEM Medium and mix thoroughly.
3.Add diluted plasmid to diluted Lipofectamine 2000 Reagent (1:1 ratio).
4.Incubate for at least 5 minutes at room temperature.
5.Digest all the 293 cells which are 90% confluent in a 10 cm (diameter) cell culture dish and suspend cells with 1 ml DMEM (10%FBS).
6.Take out 200 μl cell suspension and dilute it into 5 ml with DMEM (10%FBS).
7.Slowly drop the plasmid-lipid complex into the 5 ml cell suspension and softly mix thoroughly.
8.Plate the cell suspension into 24 well cell culture dish--1 ml each well.
9.Incubate cells for 48h at 37℃.
10.Analyze transfection efficiency in use of fluorescence microscope.
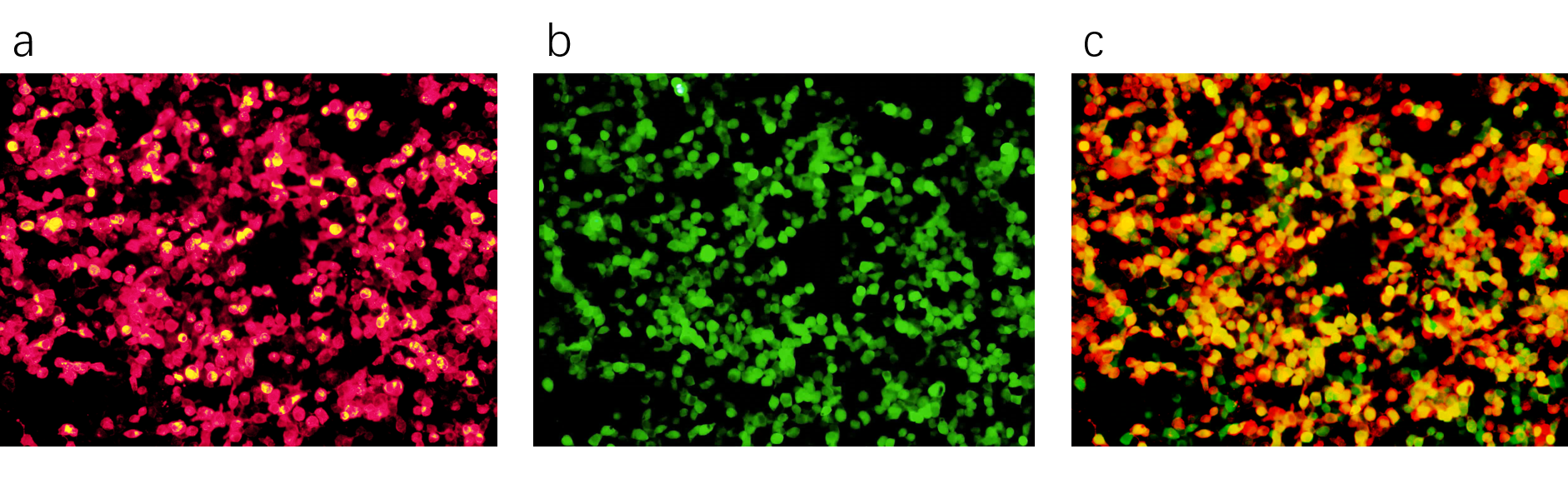
We used two different ways to stimulate Piezo1.1, and a control group is also built. The related results are shown below.
1.Yoda1
1ml 100uM Yoda1 was added to 1ml culture medium, so the final consentration of Yoda1 is 50uM. This proccess was finished under the inverted fluorescence microscope. A significant fluorescence change was observed in seconds.
Video1:Fluorescence change of cells under the stimulation of 50uM Yoda1, exposure time: 90ms, 20X objective
File:Yoda 8times.mp4
2.Shear stress
We gave the HEK293 cells the shear stress by sucking out their culture medium with a pipette. The change of fluorescence is faster and more obvious than that caused by Yoda1.
Video2:Faster fluorescence change of cells under the stimulation of shear stress, exposure time: 90ms, 20X objective
File:Force 4times.mp4
3.Control
In the control group, only GCaMP6m is transfected into the HEK293 cells. We also used 50uM of Yoda and shear stress to stimulate the cells in the control group. But no obvious change of fluorescence is observed in the control group under the condition of Yoda1 or shear stress.
Video3:Fluorescence change of cells in control group, exposure time: 90ms, 20X objective
File:Control 1.mp4
We also did a quantitative analysis of fluorescence changes. A program based on MATLAB was used to finish this quantitative analysis.
The curve showed the characteristics of rising first and then falling under the condition of Yoda1.(Figure3a) This is important to show a decreased fluorescence intensity, which is consistent with the features of Piezo1.1. The control group showed no obvious fluorescence change.(Figure3b)
These results prove that the Piezo1.1 causes calcium to flow in as we expected.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 3555
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2975
Illegal BglII site found at 7388
Illegal BamHI site found at 4125
Illegal XhoI site found at 7000
Illegal XhoI site found at 7207 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 790
Illegal NgoMIV site found at 1771
Illegal NgoMIV site found at 1798
Illegal NgoMIV site found at 2843
Illegal NgoMIV site found at 3068
Illegal NgoMIV site found at 5750
Illegal NgoMIV site found at 7312 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 877
Illegal BsaI.rc site found at 4645
Illegal BsaI.rc site found at 7471
Illegal BsaI.rc site found at 7537
Illegal SapI site found at 3741
Illegal SapI site found at 6549
Illegal SapI.rc site found at 3984
Illegal SapI.rc site found at 4690
Illegal SapI.rc site found at 5413
| None |


