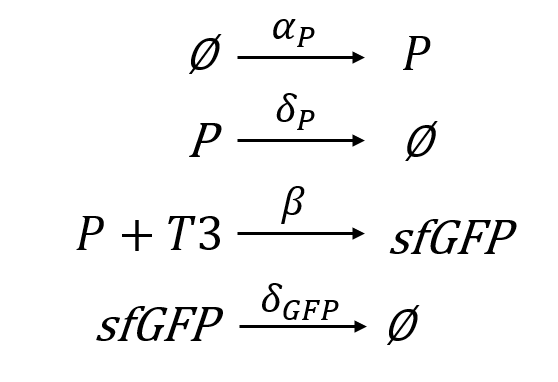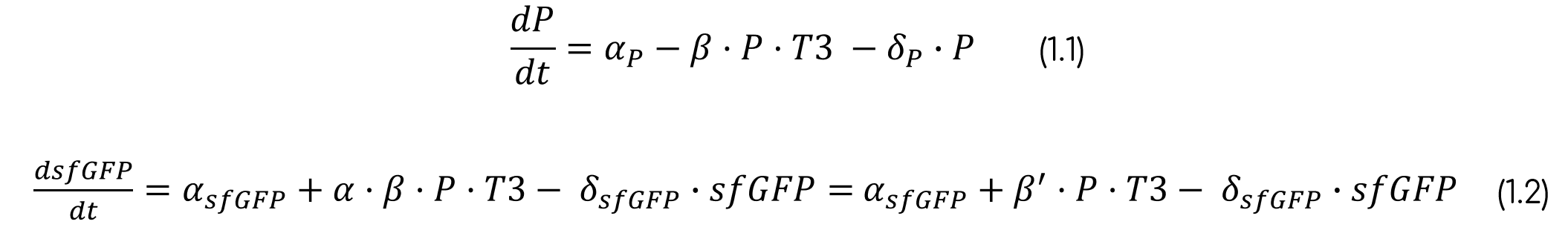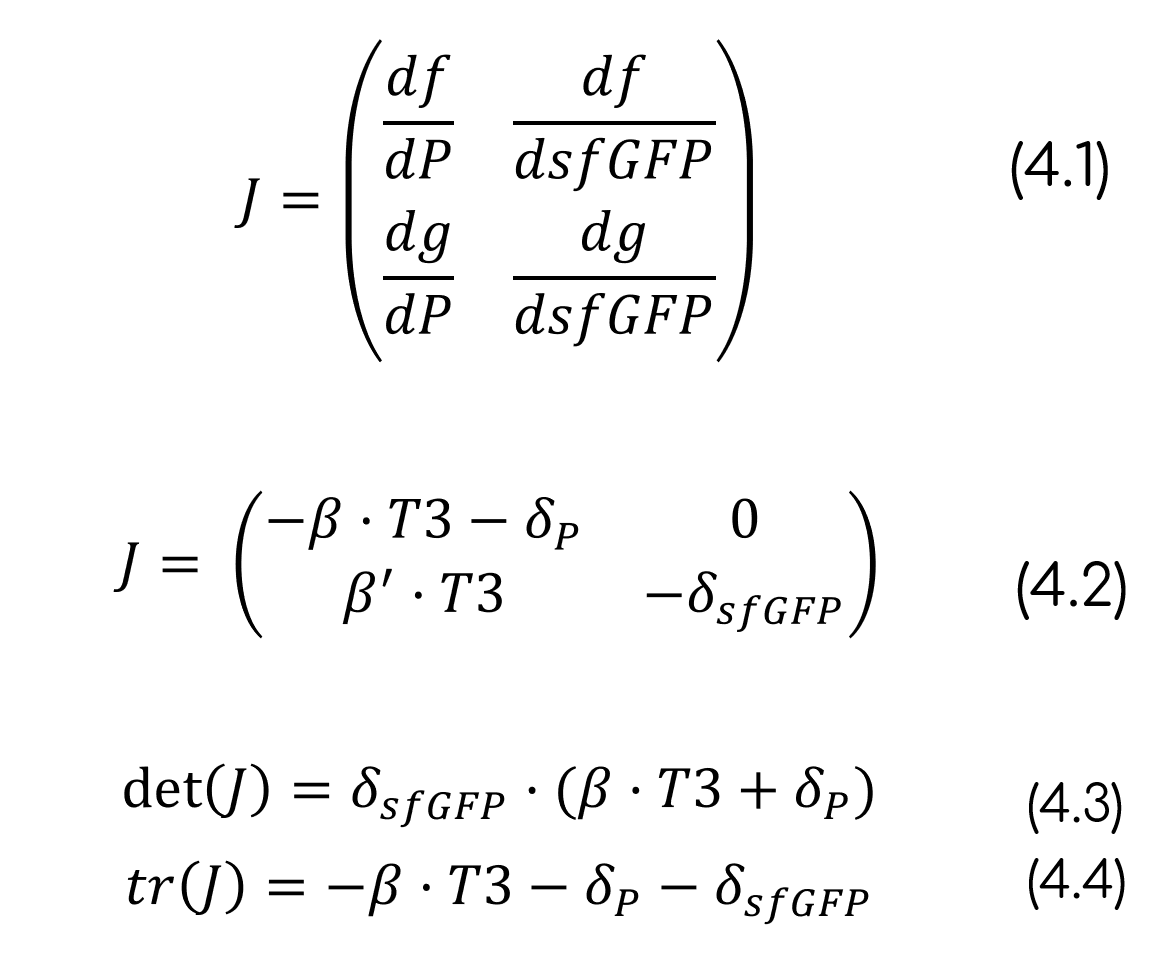Part:BBa_K3484000
Intein-mediated T3(THRB) → sfGFP
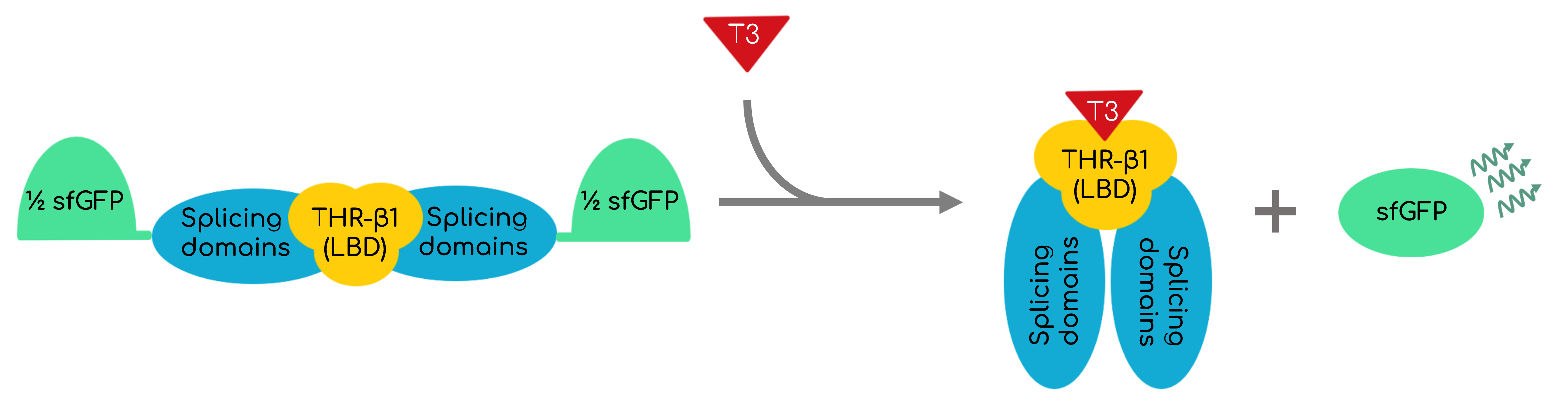
This part encodes an intein-mediated T3 biosensor. It is conformed by the Mycobacterium tuberculosis (Mtu) RecA intein, but replacing its endonuclease domain by the thyroid hormone receptor β (THR-β) ligand-binding domain (LBD). The extein (the side proteins that the intein will splice and join) is conformed by super-folder GFP (sfGFP). Being the activity of the inteins spontaneous and having a response proportional and directly dependent to the T3 in the media, a quantitative T3 biosensor that can be synthesized in bacteria and even work in cell-free systems is proportioned. Figure 1 shows in a simplified manner the functioning of the protein.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 174
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 661
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 217
Illegal SapI.rc site found at 1576
Contents
Usage and Biology
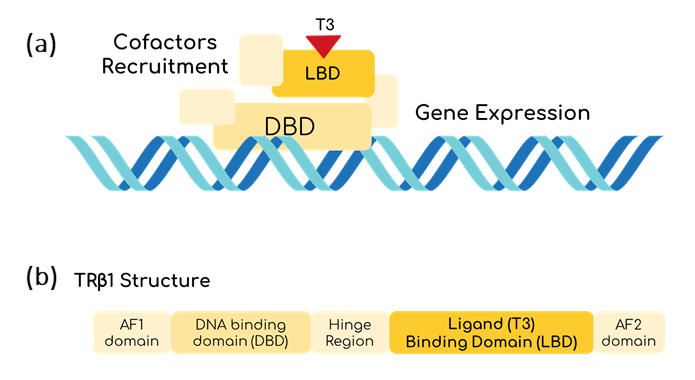
T3 could be a good marker to determine the thyroid hormonal levels of the body. To be able to sense it, THR-β seems to be useful [1]. THR-β is a nucleus protein able to bind to T3 thanks to its Ligand Binding Domain (LBD) [2]. Once this happens, THR-β binds to a specific sequence using its DNA Binding Domain (DBD). Then, it starts recruiting proteins to unpack the chromatin and to induce gene transcription (fig. 2) [3]. However, the mechanisms for gene transcription are specific for mammalian cells, so the use of inteins is proposed as an alternative to be used in bacteria.
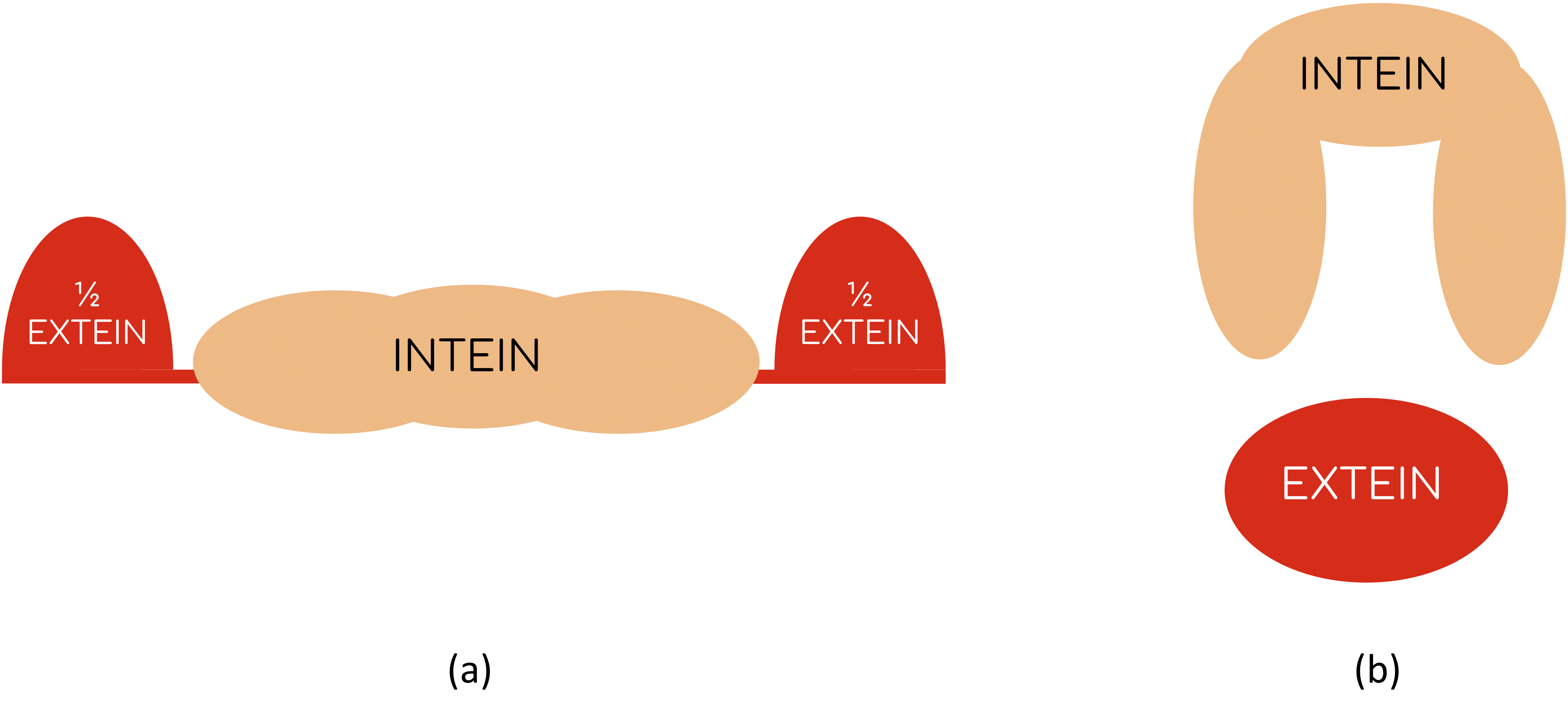
Inteins are a family of intervening proteins that carry out a post-translational event known as protein splicing: once the mechanism is activated, they are able to remove themselves from the peptide-chain joining the two remaining portions at both sides [4]. The great advantage of this process is that it is spontaneous, that means, neither external factors, not even energy sources (like ATP) are needed to splice the protein (fig. 3) [5]. A study on bacterial T3 biosensing shows that the presence of T3 can be sensed with E. coli if Mtu RecA intein is engineered with the THR-β LBD and if thymidylate synthase (essential for bacterial growth) works as the extein [6][7].
To sum up, it is shown that inteins are able to act spontaneously and that a system for detecting T3 is feasible. And it is here when it comes up the idea that having a T3 reporter that can be isolated from the cell and work in a cell-free system could be really useful, and that is what BBa_K3484000 does.
Protein complex composition
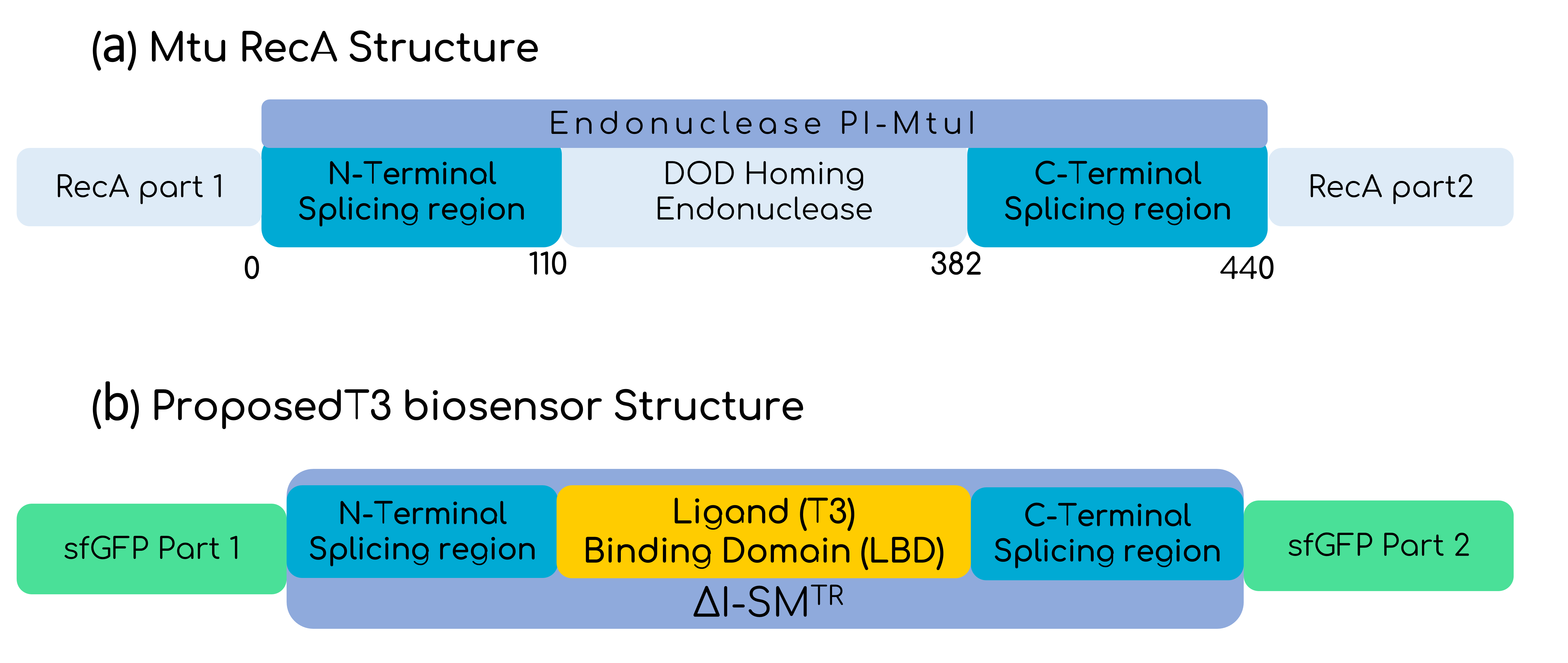
In this basic part, the intein is the optimized version of the Mtu RecA intein published by the Princeton University, which has more active splicing domains by replacing Val67 with Leu (V67L) [4]. To make the sensor sensible to T3, the homing endonuclease domain from the Endonuclease PI-MtuI from the Mtu RecA protein (110-382 residues), was substituted by the LBD of the THR-β1, only leaving the intein splicing domains from the optimized Mtu RecA intein, as shown by Gierach et al. [7]. But the great change comes when talking about the marker of the sensed T3 hormone. To make the biosensor cell-free, it is needed to think about a protein reporter that does not need of additional steps for expression, and GFP seemed to be perfect for that purpose. Moreover, as in the context of Hormonic this biosensor is intended to be coupled with electronics by the use of a photodiode, a luminescent response is ideal. Despite there are several reports on split GFPs with different inteins, fewer reports on the usage of the Mtu RecA intein with GFP are available [8][9][10]. In any case, it is important to bear in mind that the decision of the extein protein splicing point, aside from allowing an optimal fusion, needs to take into account that the C-terminal of the intein has to be followed by a Cys, Ser or Thr residue [11]. Figure 4 shows how finally the construct looks like. Furthermore, as Hormonic is a diagnostic and therapeutic tool, it is needed that the signal is rapidly generated and that is why sfGFP (split between 70th-71st residues) is used instead of other variants that have more reports on its usage with the Mtu RecA intein, but that are slower at expressing [12]. Given the propensity of the intein-GFP complex to form inclusion bodies, as Gangopadhyay et al. had shown, we added the solubility tag Fh8 at the N-terminal of the sfGFP, considering its short length and its high solubility power [13]. Finally, we added the FLAGx3 tag at the C-terminal of the sfGFP for immunodetection purposes.
Functionalization
| super-folder GFP expression | Protein domain | BBa_K3484000 |
| Transcriptional unit | BBa_K3484001 | |
| enhanced GFP expression | Protein domain | BBa_K3484002 |
| Transcriptional unit | BBa_K3484003 |
Characterization
Modelling
Considering the interactions found on the T3 sensor cell previously described and shown in figure 1, when T3 binds to the receptor, the intein splices, reconstructing the sfGFP, thus generating fluorescence that can be measured. We must consider that this reconstruction is not 100% reliable, thus leading also to non-functional fluorescent proteins [14]. Moreover, the construct is preceded by a constitutive promoter, therefore, the intein is produced constitutively in the cell, although there is only fluorescence when T3 is present in the media and the intein domain is activated.
From these interactions we can develop 4 reactions (fig. 5) which are the basis for our T3 sensor cell model (the parameters are defined in tables 2 and 3).
| Parameters | Description | Units | Fitted values | Normalized fitted values |
| αP | Production rate of P | (a. u.)/time | 7.14 · 103 | 5.89 · 103 |
| δP | Degradation rate of P | 1/time | 4.85 · 10-2 | 5.03 · 103 |
| β | Production rate of sfGFP | 1/(time ·(a. u.)) | 8.74 · 103 | 9.07 · 103 |
| αsfGFP | Basal Production rate of sfGFP | RFU/time | 4.00 · 103 | 2.63 · 103 |
| δsfGFP | Degradation rate of sfGFP | 1/time | 5.14 | 6.28 · 103 |
| β' | Production rate of functional sfGFP. | 1/time(a.u.) | 7.14 · 103 | 5.89 · 103 |
| Variables | Units |
| P | Arbitraty units (a. u.) |
| T3 | Molar concentration (M) |
| GFP | Relative fluorescence units (RFU) |
Due to the experimental results, we must consider that there is a basality in the production of sfGFP. Moreover, we are taking into account that the variable P always refers to the whole intein construct, including the split sfGFP at both intein extremes. Therefore, when sfGFP is generated, this construct is degraded. This makes sense as the residual construct is non-functional, since it has already spliced the sfGFP. Furthermore, as said before, it is necessary to multiply the production of sfGFP for a rate, as the reconstruction can lead to non-functional sfGFP. The final ODE system and its parameters and variables can be seen below (eq. 1, tab. 2-3). The parameters were fitted with the fluorescence normalized from 0 to 1 and non-normalized.
From the ODE system (eq. 1) a transfer function can be derived if P and sfGFP are on a steady-state (dy/dx=0). This transfer function (eq. 2) allows us to connect the concentration of T3 in the media to the fluorescence that the sensor cells are emitting. It results on a Hill function with a low Hill coefficient (n=1), thus meaning that the sensor will not be hypersensitive and it will have a large dynamic range where it can sense the T3 concentration.
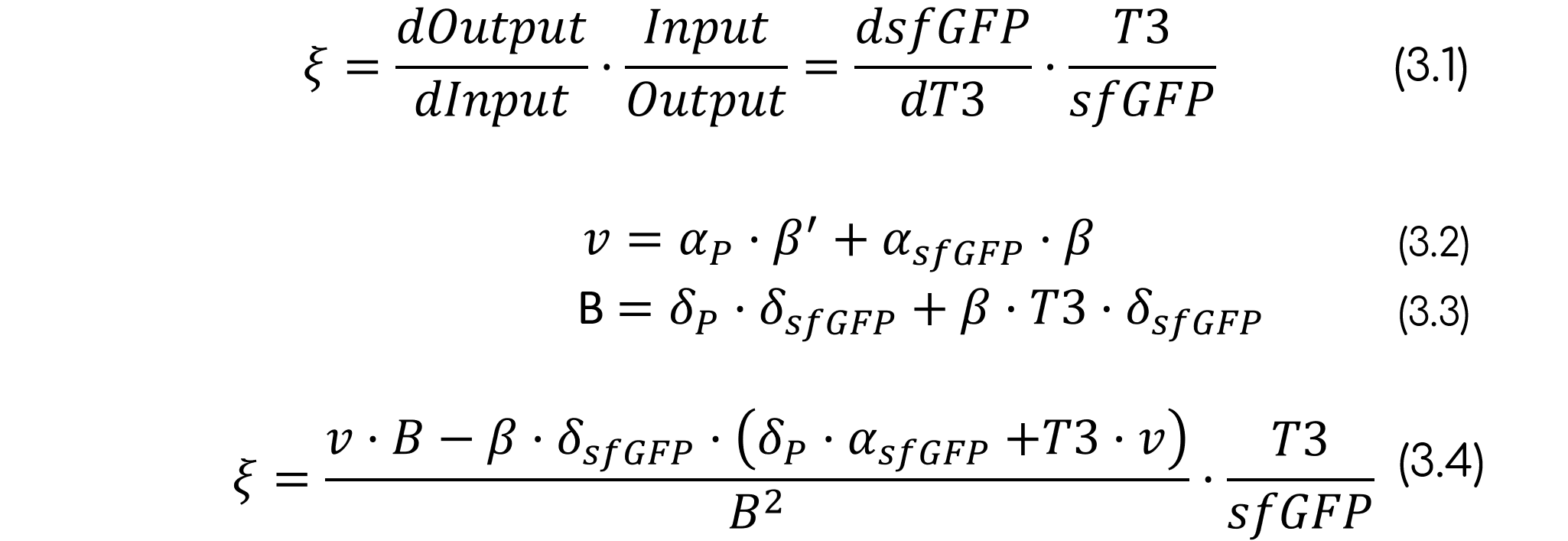
The sensitivity of the system is a good parameter to determine the ability to detect and react to changes in the measured physical quantity. In our case, it will be representing the minimum change in T3 concentration (M) that generates a change in the fluorescence, expressed in relative fluorescence units (RFU). In order to calculate the sensitivity, we used its mathematical definition (eq. 3), which says that the sensitivity is the derivative normalized to the input and output.
A sensitivity around 0 and 1 (E=0.2107) is obtained, which means that the sensor is neither robust (which would mean that the biosensor cannot react to different T3 concentrations), nor hypersensitive (which would mean that the biosensor overreacts to minimal changes in the T3 concentration, thus shrinking the dynamic range), respectively.
To analyze the stability of the system at the steady-state we can use the Jacobian of the ODE system (J). From the Jacobian we can compute the trace [tr(J)] and the determinant [det(J)] so that we can analyze the stability of the system. Calculations of the different parameters have been solved for Equation 4.
Since the determinant is positive and the trace is negative (eq. 4), we can affirm that the system has a stable steady-state [15]. This means that even if the system is far from the steady-state, for example when we include T3 to the media and sfGFP has not yet been formed, the system will evolve towards the steady-state, thus sensing the T3 concentration.
Experimental methodology
In order to characterize the functionality of BBa_K3484000, it was necessary to create a transcriptional unit with the BBa_K880005 Promoter + Ribosome Binding Site and BBa_B0014 double terminator. This resulting construct is the composite part: BBa_K3484001. BBa_K3484001 in high copy pUC backbone was transformed in E. coli BL21 (DE3) and plate-reader experiments were set up to sense the amount of fluorescence produced by our biosensor in different T3 concentrations. BL21 (DE3) containing the BBa_K3484001 plasmid and no plasmid (negative control) were grown overnight in Luria Broth (LB) media. The next day, 1 part cells for 30 fresh LB media were inoculated and grown for 3 to 4 hours. The construct cells and the negative control cells were induced with different concentrations of T3: 0M, 1pM, 10pM, 100pM, 1nM, 10nM, 100nM, 500nM, 1uM, 5uM, 10uM and 100uM plus blank controls. Fluorescence intensity and optical density (OD) at wave-length 600 nm were analysed every 30 minutes over 24 hours with a plate-reader (tab. 4).
| Parameters | Value |
| Plate-Reader model | Synergy HTX |
| Plate type | Thermo Fischer 96-well microplates black-walled clear bottom |
| Cell medium | LB |
| Time | 24 hours |
| Shake | Linear: Continuous, Frequency: 567 rpm (3mm) |
| Temperature | 37 ºC |
| Gain | 50 |
| Optical Density (OD) measurement (absorbance) | 660 nm |
| sfGFP excitation wavelength | 485 nm |
| sfGFP emission wavelength | 528 nm |
Results
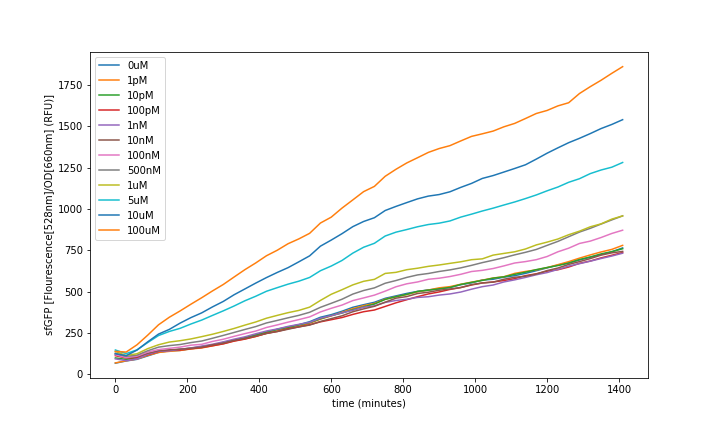
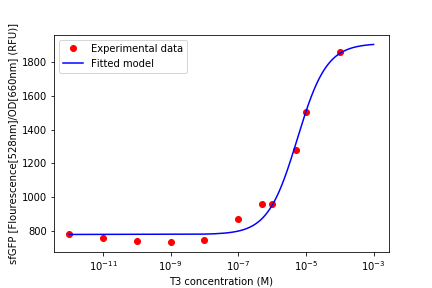
Fluorescence was obtained by subtracting the blank LB fluorescence and normalizing to the OD (fig. 6). All the values were obtained from the mean of the 3 same condition wells. Then it was analysed at steady-state to generate the transfer functions (fig. 8).
IMT3_sfGFP_pUC-Amp fluorescence over the time showed an increase proportional to the concentration of T3 in the media (fig. 6).
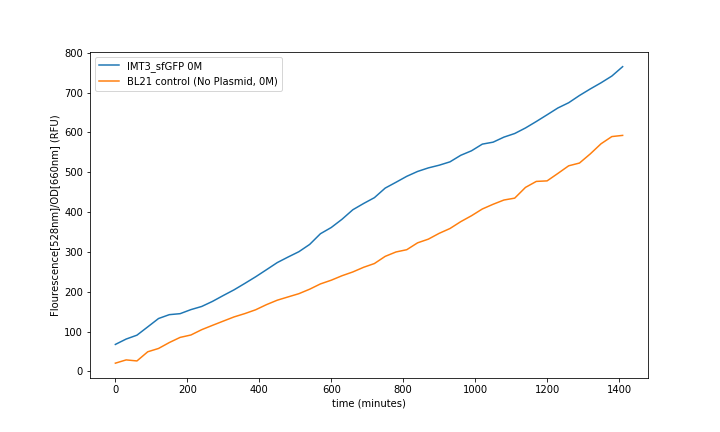
Control of the BL21 without plasmid and BL21 with plasmid and no T3 induction showed the leaking fluorescence of the part (fig. 7).
We could observe that the sensor response had a dynamic range of: 10-07 to 10-04 M of T3, as can be seen above (fig. 8). The resulting data was used to fit the model seen above and we could see that it described the sensor’s dynamics.
References
[1] Abdalla, S. M., & Bianco, A. C. (2014). Defending plasma T3 is a biological priority. Clinical Endocrinology, 81(5), 633–641. DOI:10.1111/cen.12538
[2] Yen, P. M. (2001). Physiological and Molecular Basis of Thyroid Hormone Action. Physiological Reviews, 81(3), 1097–1142. DOI:10.1152/physrev.2001.81.3.1097
[3] Flamant, F., Cheng, S.-Y., Hollenberg, A. N., Moeller, L. C., Samarut, J., Wondisford, F. E., Refetoff, S., et Al. (2017). Thyroid Hormone Signaling Pathways: Time for a More Precise Nomenclature. Endocrinology, 158(7), 2052–2057. DOI:10.1210/en.2017-00250
[4] Wood, D. W., Wu, W., Belfort, G., Derbyshire, V., & Belfort, M. (1999). A genetic system yields self-cleaving inteins for bioseparations. Nature Biotechnology, 17(9), 889–892. DOI:10.1038/12879
[5] Shah, N. H., & Muir, T. W. (2014). Inteins: nature’s gift to protein chemists. Chem. Sci., 5(2), 446–461. DOI:10.1039/c3sc52951g
[6] Skretas, G. (2005). Regulation of protein activity with small-molecule-controlled inteins. Protein Science, 14(2), 523–532. DOI:10.1110/ps.04996905
[7] Gierach, I., Li, J., Wu, W.-Y., Grover, G. J., & Wood, D. W. (2012). Bacterial biosensors for screening isoform-selective ligands for human thyroid receptors α-1 and β-1. FEBS Open Bio, 2(1), 247–253. doi:10.1016/j.fob.2012.08.002
[8] Ozawa, T., Takeuchi, M., Kaihara, A., Sato, M., & Umezawa, Y. (2001). Protein Splicing-Based Reconstitution of Split Green Fluorescent Protein for Monitoring Protein−Protein Interactions in Bacteria: Improved Sensitivity and Reduced Screening Time. Analytical Chemistry, 73(24), 5866–5874. DOI:10.1021/ac010717k
[9] Ozawa, T., & Umezawa, Y. (2001). Detection of protein-protein interactions in vivo based on protein splicing. Current Opinion in Chemical Biology, 5(5), 578–583. DOI:10.1016/s1367-5931(00)00244-1
[10] Ozawa, T., Nogami, S., Sato, M., Ohya, Y., & Umezawa, Y. (2000). A Fluorescent Indicator for Detecting Protein−Protein Interactions in Vivo Based on Protein Splicing. Analytical Chemistry, 72(21), 5151–5157. DOI:10.1021/ac000617z
[11] Tornabene, P., Trapani, I., Minopoli, R., Centrulo, M., Lupo, M., de Simone, S., Auricchio, A., et Al. (2019). Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Science Translational Medicine, 11(492), eaav4523. DOI:10.1126/scitranslmed.aav4523
[12] Gangopadhyay, J. P., Jiang, S., & Paulus, H. (2003). An in Vitro Screening System for Protein Splicing Inhibitors Based on Green Fluorescent Protein as an Indicator. Analytical Chemistry, 75(10), 2456–2462. DOI:10.1021/ac020756b
[13] Costa, S., Almeida, A., Castro, A., & Domingues, L. (2014). Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Frontiers in Microbiology, 5. DOI:10.3389/fmicb.2014.00063
[14] Gangopadhyay, J. P., Jiang, S., & Paulus, H. (2003). An in Vitro Screening System for Protein Splicing Inhibitors Based on Green Fluorescent Protein as an Indicator. Analytical Chemistry, 75(10), 2456–2462. DOI:10.1021/ac020756b
[15] Layek, G.C. (2015). “Stability Theory” in An Introduction to Dynamical Systems and Chaos 1rst ed. Springer, pp. 129-158. DOI:10.1007/978-81-322-2556-0
//chassis/prokaryote/ecoli
//function/reporter/fluorescence
| None |