Part:BBa_K3174008
SYFP2 coding sequence with IS hotspot removed
For more information on how this part was conceived and designed, please visit the design page.
This is a redesigned version of the Super Yellow Fluorescent Protein 2 (Part:BBa_K864100) that has had a region that was prone to IS10 insertion element mutations altered. This removal of a mutation hotspot resulted in the expression of SYFP2 in TOP10 E. coli cells being maintained for a longer period of time than the original sequence.
Usage and Biology
This part codes for the bright yellow fluorescent protein SYFP2, which has an excitation peak of 515 nm and an emission peak of 527 nm.
Testing and Measurement
Cultures of TOP10 E. coli were transformed with either the original SYFP2 plasmid or the redesign and propogated over four days. Each day a new culture was created using a 1:1000 dilution of the previous day's culture and allowed to grow for 24 hours at 37 C. Fluorescence was measured daily using flow cytometry. The data showed that while the original plasmid stopped expressing fluorescence after one day, the redesigned plasmid retained expression for up to four days.
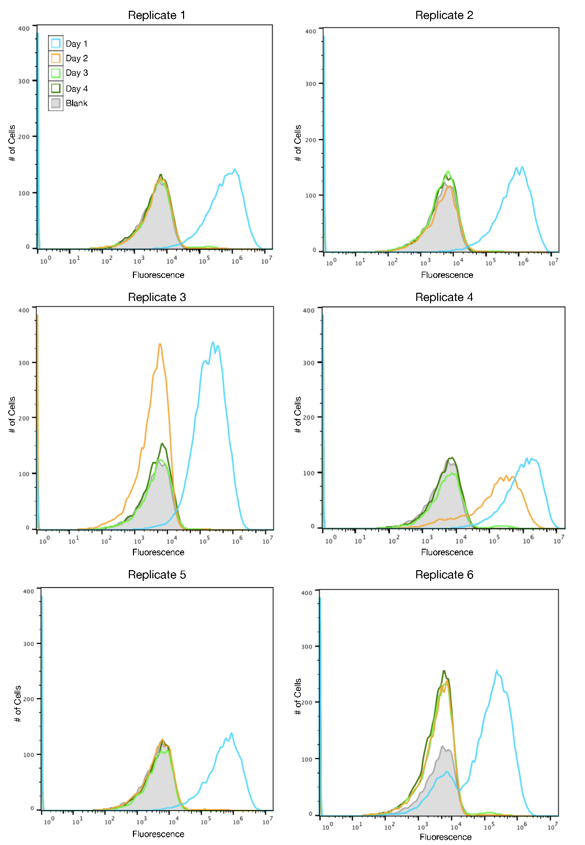
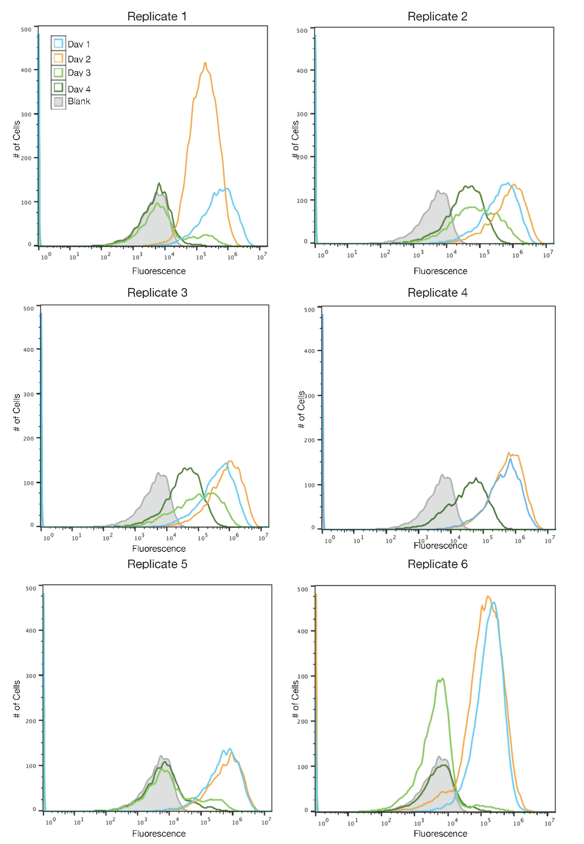
This data indicated of functional improvement of the redesigned SYFP2 sequence over the original. While the original sequence mutated out of culture within a day, the redesign lasted up to four days of propagation in TOP10 cells. This improvement is particularly useful as TOP10 is a common competent cell strain used in synthetic biology research. By making the SYFP2 sequence more resilient to IS10 mutations, it can be used more effectively in TOP10 cells which are known to carry IS elements in their genomes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |
