Part:BBa_K2549042
NLS-TEVpN-CfaN
This part is one of the downstream elements of our amplifier. It is constructed by fusing NLS (Part:BBa_K2549054), TEVpN (Part:BBa_K2549014) and CfaN (Part:BBa_K2549009), from N terminal to C terminal. NLS is a short nuclear location sequence from SV40 large T antigen. TEVpN is the N-terminal fragment of TEV protease. TEV protease is one of the best-characterized enzymes of the viral proteases which have more stringent sequence specificity. CfaN is the N-terminal fragment of Cfa which is a consensus sequence from an alignment of 73 naturally occurring DnaE inteins that are predicted to have fast splicing rates[1]. When coexpressed with CfaC-TEVpC (Part:BBa_K2549043) in the same cell, two fusion proteins can form and the site-specific cleavage can be realized.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 337
Biology
Please visit http://2018.igem.org/Team:Fudan/Results and http://2018.igem.org/Team:Fudan/Measurement to check how we use this.
split TEV protease
MJ Rossner et al have demonstrated a biological assay termed split TEV. They engineered inactive fragments of the NIa protease from the tobacco etch virus (TEV protease) that regain activity only when coexpressed as fusion constructs with interacting proteins[2].
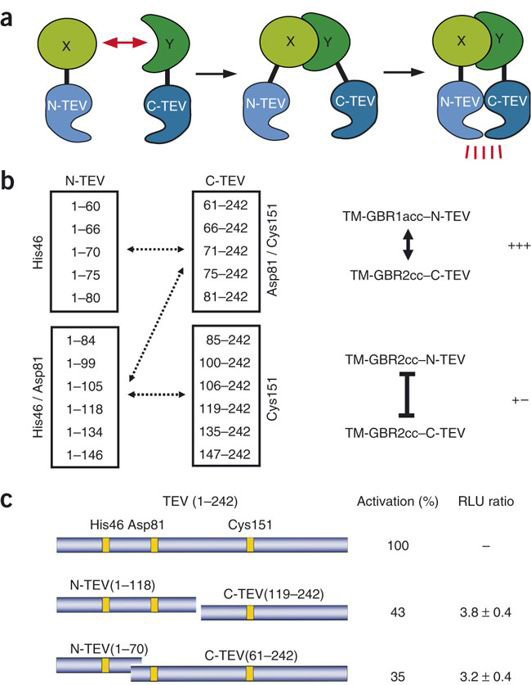
Thus in our split TEV protease assay, we chose site between 118 amino acid and 119 amino acid of TEV protease as the split site.
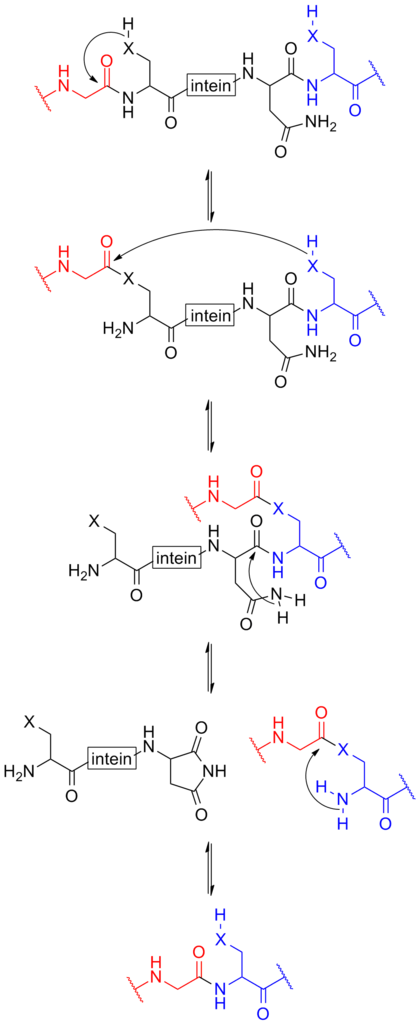
intein-based protein splicing
Schematic below provides a mechanism of how the intein function[3].
For more details about intein-based protein splicing, please refer to our CfaN (Part:BBa_K2549009) and CfaC (Part:BBa_K2549010).
References
- ↑ Design of a Split Intein with Exceptional Protein Splicing Activity. Stevens AJ, Brown ZZ, Shah NH, ..., Cowburn D, Muir TW. J Am Chem Soc, 2016 Feb;138(7):2162-5 PMID: 26854538; DOI: 10.1021/jacs.5b13528
- ↑ Monitoring regulated protein-protein interactions using split TEV. Wehr MC, Laage R, Bolz U, ..., Nave KA, Rossner MJ. Nat Methods, 2006 Dec;3(12):985-93 PMID: 17072307; DOI: 10.1038/nmeth967
- ↑ https://en.wikipedia.org/wiki/Intein
//proteindomain/internal/special
//proteindomain/localization
| None |