Part:BBa_K2017007
35s:5'+ Ga20ox consense + SAGTI-Luciferase + Tnos
Modul of the gRNA testing system created by Valencia UPV 2016 team. This system aims to test the functionality and efficiency of a chosen gRNA to target a particular gene. This system is modular, as the gene inserted can be chosen by the user, only taking into account the required overhangs (5'-AATG-3' and 5'-TTCG-3')
It includes a promoter 35s with a 5' region (BBa_K2017002), 20 nucleotides of the Ga20ox consensus gene of Oryza sativa (rice), the reporter luciferase with SAGTI linker and the terminator Tnos.
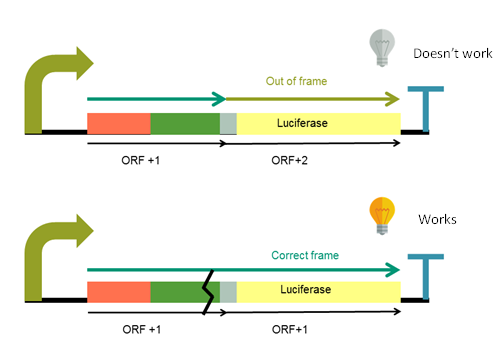
Luciferase is out of its reading frame due to a nucleotide added upstream to the linker, and the initial ATG has been removed. This means that when the luciferase is translated, it will not be functional. To make it functional, it is necessary to make indels in the Ga20ox fragment that put the luciferase in the correct reading frame. These indels can be made using the CRISPR/Cas9 system, which is the aim of this device.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1050
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1070
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 2765
Illegal BsaI.rc site found at 3052
Illegal SapI.rc site found at 1911
Usage and Biology
The gRNA testing system is a modular and standard Phytobrick device. It is composed by modular parts of the Parts Registry. A plant promoter (35s), a fragment of the targeted gene chosen by the user using our [http://2016.igem.org/Team:Valencia_UPV/Software Data Processing Software], a linker with luciferase (BBa_K2017003, BBa_K2017004, BBa_K2017005, BBa_K2017006) and a plant terminator (Tnos). The objective of the gRNA testing system is to test a target of a chosen gene in a model plant as N. benthamiana, to know if it would work or not in the plant that wants to be modified. This is made to save time and test a gRNA in a few days, instead of waiting days to know if the editing has worked in the plant to modify.
This device is introduced in the plant along with the corresponding gRNA and Cas9 construction. If the gRNA leads Cas9 to the target in the testing system and mutations are produced, bioluminescence signal will be detected through a luciferase assay. The main advantages of this kind of assays are its straightforward protocol (the analysis of each sample only requires a few minutes) its enormous sensitivity and its accuracy on the obtained results. Thus, allowing us to obtain quantitative results.
The system is designed in order to be in OFF state until the double strand break is produced by the Cas9 and subsequent repair is produced. When Cas9 cuts indels appear, luciferase open reading frame returns to the correct one and system turns ON (Figure 1).
Leaf samples are assayed for the presence of the reporter gene by directly measuring the enzymatic activity of luciferase enzyme in the presence of known concentration of luciferin.
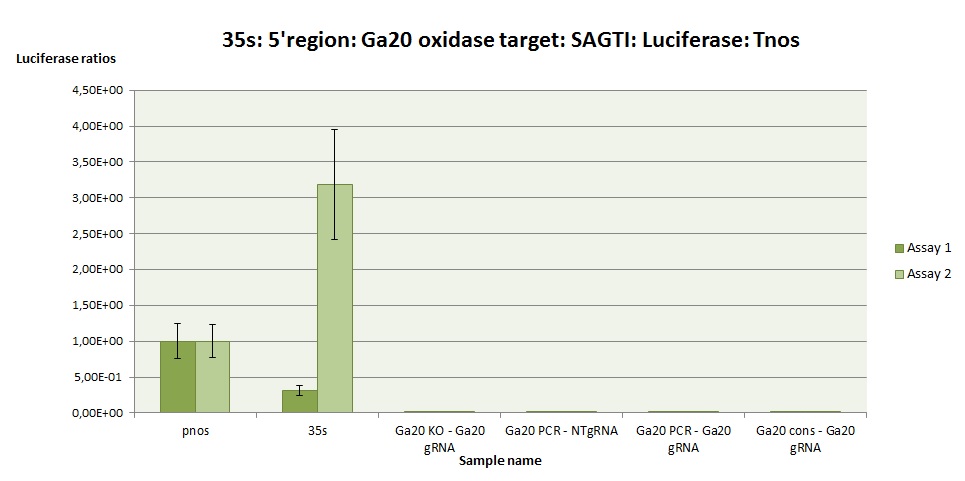
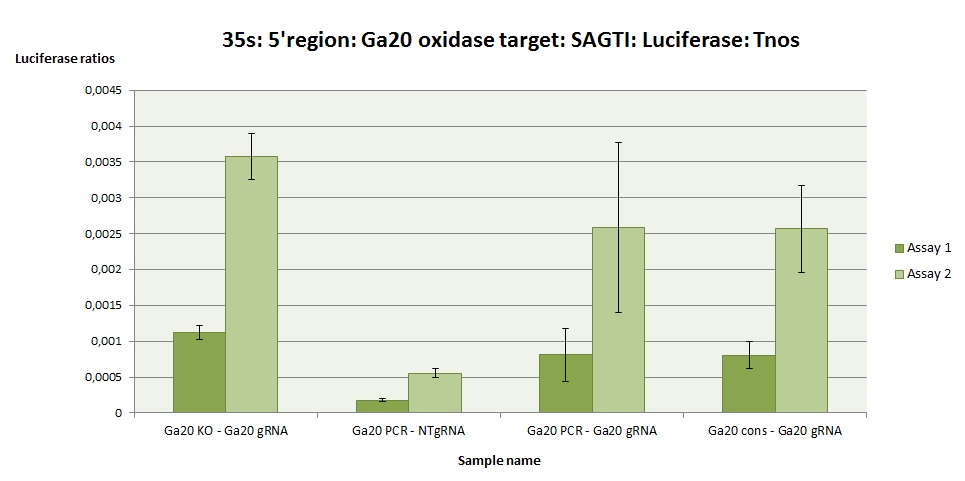
The Ga20-KO testing system contains the target gene with one deletion, to mimic the testing system edited by Cas9. It is the positive control (check BBa_K2017004). The results in Ga20-KO show that the gRNA testing system would work when edited by CRISPR/Cas9. pNOS and 35s express luciferase constitutively, and serve as standard controls for luciferase essays in plant.
In this case it is tested the Ga20ox gene of rice (Oryza sativa) (Figure 2 and 3), with the objective to know the efficiency of the chosen target for editing this gene in rice. The experiment is fully repeated twice (assay 1 and 2).
The Ga20-PCR is a testing system with the target amplified directly from the original plant. The Ga20-cons is a target the consensus sequence of the gene, obtained from the NCBI database. As it can be seen in the figures, the tested gRNA testing systems (Ga20-PCR and Ga20-cons) express higher levels of luciferase than the negative control (NT, without gRNA), meaning that the testing system can be turned on when the Cas9 acts on the target.
//chassis/eukaryote/nbenthamiana
//function/crispr
| None |