Difference between revisions of "Part:BBa K1949101"
| Line 21: | Line 21: | ||
<span style="margin-left: 10px;">We constructed | <span style="margin-left: 10px;">We constructed | ||
| − | + | <i>E. coli</i> A: Pcon - <i>rbs</i> - <i>gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | + | <i>E. coli</i> B: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3). | |
=====result===== | =====result===== | ||
| Line 32: | Line 32: | ||
====Ⅱ.<i>mazEF</i> System Assay ~Stop & GO~==== | ====Ⅱ.<i>mazEF</i> System Assay ~Stop & GO~==== | ||
| − | <span style="margin-left: 10px;">The | + | <span style="margin-left: 10px;">The most attractive feature of the <i>mazEF</i> system is that cytotoxity of a toxin protein (MazF) is determined by the stoichiometric ratio of a toxin protein to a corresponding antitoxin protein (MazE) in cells. The story of Snow White involves sleep and resuscitate from sleep, and thus, we think that this feature is useful for our project.<br> |
<span style="margin-left: 10px;">We constructed | <span style="margin-left: 10px;">We constructed | ||
| − | + | <i>E. coli</i> C: PBAD - <i>rbs</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | + | <i>E. coli</i> A: Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | + | <i>E. coli</i> D: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs - mazE</i> (pSB3K3) | |
| − | + | <i>E. coli</i> B: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3). | |
=====result===== | =====result===== | ||
| − | <span style="margin-left: 10px;"> | + | <span style="margin-left: 10px;">As shown in Fig. 2, when the mazE expression was induced 2 h after the mazF expression, cell growth resumed gradually. Similarly, the RFU of GFP also increased mazF expression, and about 8 h later, cell growth resumed. Similarly, the RFU of GFP was also resumed. Before starting the experiments, we anticipated that recovery from the growth inhibition by mazF would be observed immediately after the induction of mazE expression. However, it took 8 h for us to observe the recovery. The reason is unclear, but the prior expression of mazF might damage the cells and it might take time to repair. Alternatively, the mazF expression might lead cells to the dormant state from which cells can hardly escape. We assume that this feature is advantageous for our Snow White project, because we can prolong the sleeping time of Snow White and take sufficient time for Prince to find Snow White. As a conclusion, the “Stop and Go” experiment was successful. |
| + | |||
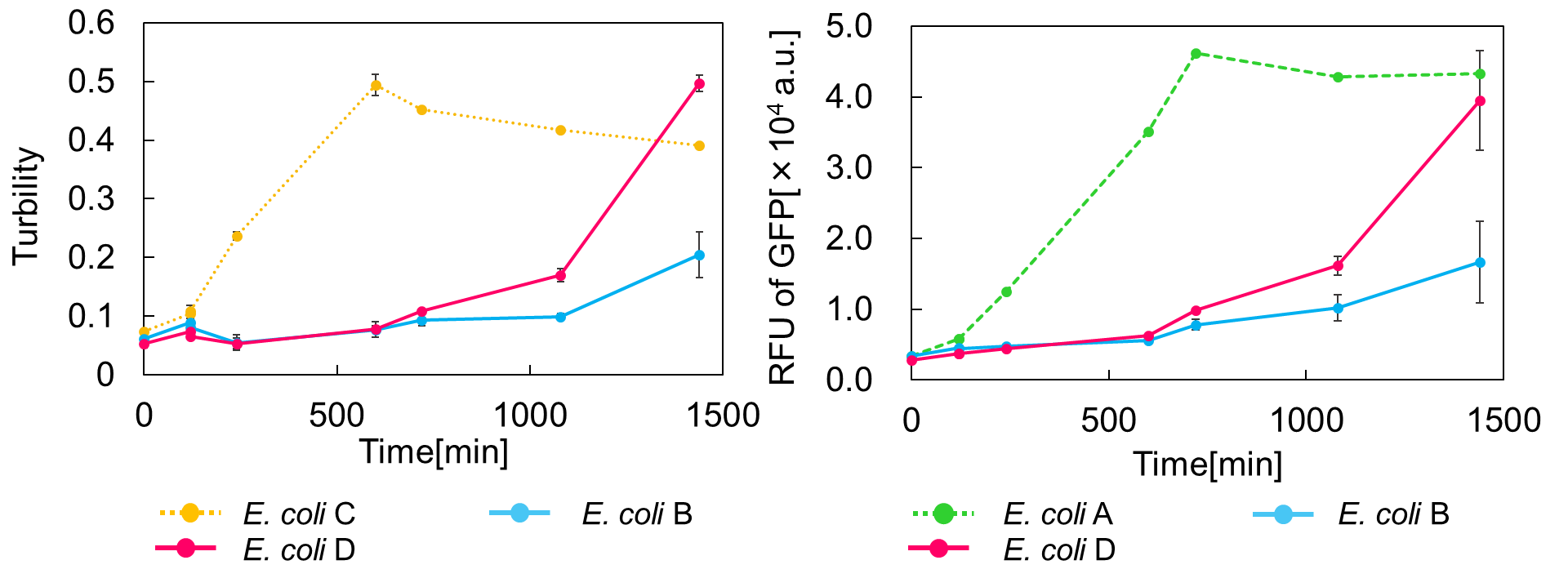
[[Image:Stop and Go.png|thumb|center|600px|Fig. 2. Time vs Turbidity (left), Time vs RFU of GFP (right)]]<br> | [[Image:Stop and Go.png|thumb|center|600px|Fig. 2. Time vs Turbidity (left), Time vs RFU of GFP (right)]]<br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
====Ⅲ.<i>mazEF</i> System Assay ~Go & Stop~==== | ====Ⅲ.<i>mazEF</i> System Assay ~Go & Stop~==== | ||
| − | <span style="margin-left: 10px;"> | + | <span style="margin-left: 10px;">In experimentⅡ, a toxin inhibits cell growth, and an antitoxin resuscitates it. However, what will happen when a toxin is expressed after the constitutive expression of antitoxin? Therefore, in the experiments of this section, we conducted a reciprocal experiment and named this experiment "Go & Stop".<br> |
<span style="margin-left: 10px;">We constructed | <span style="margin-left: 10px;">We constructed | ||
| − | + | <i>E. coli</i> C: PBAD - <i>rbs</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | + | <i>E. coli</i> A: Pcon - <i>rbs - gfp</i> (pSB6A1), Plac - <i>rbs</i> (pSB3K3) | |
| − | + | <i>E. coli</i> G: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Pcon - <i>rbs</i>(weak) - <i>mazE</i> (pSB3K3) | |
| − | + | <i>E. coli</i> F: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), Pcon - <i>rbs - mazE</i> (pSB3K3) | |
| − | + | <i>E. coli</i> E: PBAD - <i>rbs - mazF - tt</i> - Pcon - <i>rbs - gfp</i> (pSB6A1), vector (pSB3K3). | |
=====result===== | =====result===== | ||
| − | <span style="margin-left: 10px;"><i>E. coli</i> | + | <span style="margin-left: 10px;">Turbidity of <i>E. coli</i> G stopped earlier than that of <i>E. coli</i> F (Fig. 3), indicating that cellular <i>mazE</i> amount over MazF amount affects cytotoxity of MazF. In other words, the cytotoxity of MazF is determined stoichiometrically. Similarly, final RFU of <i>E. coli</i> G was lower than that of <i>E. coli</i> F. |
| + | <span style="margin-left: 10px;">From the results of ExperimentⅠ and ExperimentⅡ, it is expected that <i>mazEF</i> can repeatedly control cell growth. | ||
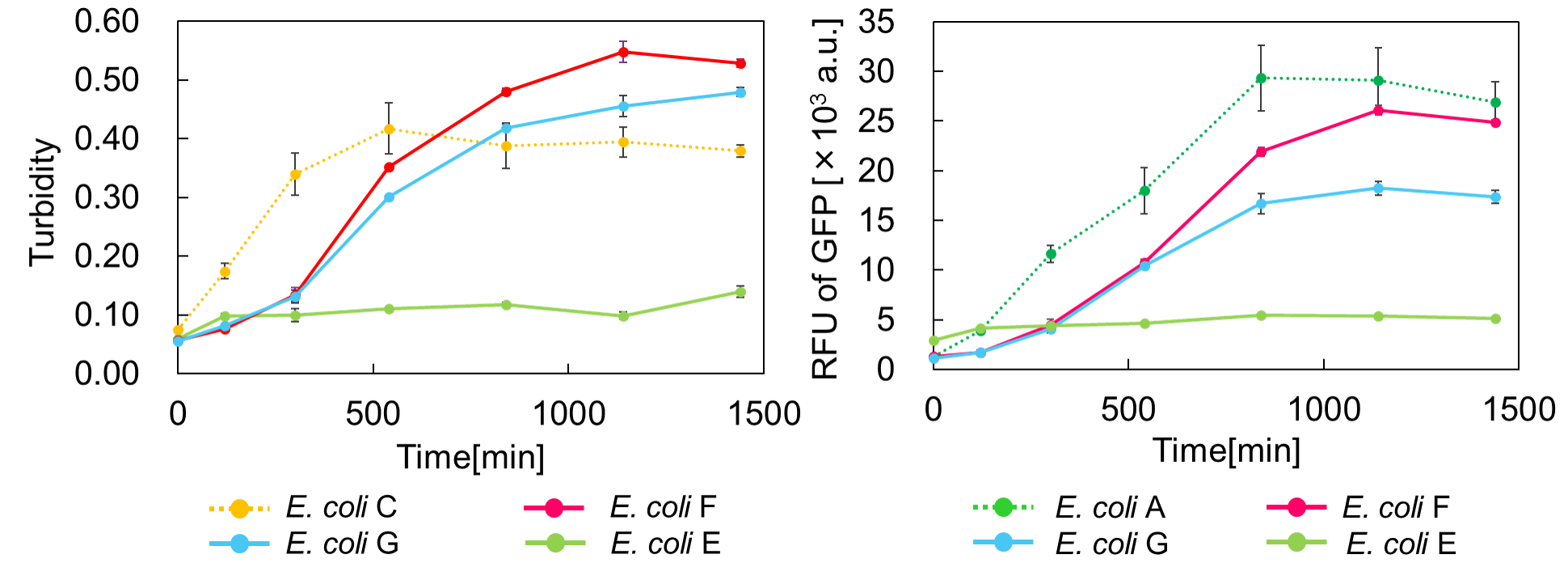
| + | [[Image:GO & STOP.png|thumb|center|600px|Fig. 3. Time vs Turbidity (left), Time vs RFU of GFP (right)]]<br> | ||
| + | |||
| + | <span style="margin-left: 10px;">Calculation of the change of RFU of GFP / Turbidity per unit time (translation efficiency) indicates that the expression level of <i>mazE</i> correlated with the translation efficiency(Fig. 4). | ||
[[Image:GO & STOP.png|thumb|center|600px|Fig. 3. Time vs Turbidity (left), Time vs RFU of GFP (right)]]<br> | [[Image:GO & STOP.png|thumb|center|600px|Fig. 3. Time vs Turbidity (left), Time vs RFU of GFP (right)]]<br> | ||
| − | |||
| − | |||
====Ⅳ.Control of Cell Growth==== | ====Ⅳ.Control of Cell Growth==== | ||
Revision as of 19:15, 19 October 2016
PBAD-rbs-mazF
We designed parts(BBa_K1949100, BBa_K1949101, BBa_K1949102, BBa_K1949103, BBa_K1949104) to characterize mazE(BBa_K1096001) and mazF(BBa_K1096002).
Characterize
We performed experiment to work our final genetic circuits and characterized mazE and mazF.
This experiment consists of the four parts below.
Ⅰ.Adjustment of MazF Expression
Ⅱ.mazEF System Assay ~Stop & GO~
Ⅲ.mazEF System Assay ~Go & Stop~
Ⅳ.Control of Cell Growth
Ⅰ.Adjustment of MazF Expression
In order to control cell growth as we desire using the mazEF system, it is necessary to adjust the expression level of mazF. It has been reported that MazF has very strong ability to inhibit cell growth and that mazE expression can not recover it when mazF is expressed at high level[1]. Therefore, we here explored the relationship between concentration of the expression inducer for mazF (arabinose in this experiment) and expression level of it; such information is important for operating our final genetic circuits properly.
We constructed
E. coli A: Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
E. coli B: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3).
result
It was found that cell growth of E. coli B was inhibited under arabinose (inducer for mazF) concentrations of over 0.02% (Fig. 1). Interestingly, at arabinose concentration of 0.2%, GFP fluorescence intensity fell markedly, regardless of mazF expression, suggesting that high arabinose concentrations may inhibit gfp expression or prevent GFP from exerting fluorescence. Taken together, it is concluded that the concentration of arabinose should be 0.02%.
Ⅱ.mazEF System Assay ~Stop & GO~
The most attractive feature of the mazEF system is that cytotoxity of a toxin protein (MazF) is determined by the stoichiometric ratio of a toxin protein to a corresponding antitoxin protein (MazE) in cells. The story of Snow White involves sleep and resuscitate from sleep, and thus, we think that this feature is useful for our project.
We constructed
E. coli C: PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
E. coli A: Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
E. coli D: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs - mazE (pSB3K3)
E. coli B: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3).
result
As shown in Fig. 2, when the mazE expression was induced 2 h after the mazF expression, cell growth resumed gradually. Similarly, the RFU of GFP also increased mazF expression, and about 8 h later, cell growth resumed. Similarly, the RFU of GFP was also resumed. Before starting the experiments, we anticipated that recovery from the growth inhibition by mazF would be observed immediately after the induction of mazE expression. However, it took 8 h for us to observe the recovery. The reason is unclear, but the prior expression of mazF might damage the cells and it might take time to repair. Alternatively, the mazF expression might lead cells to the dormant state from which cells can hardly escape. We assume that this feature is advantageous for our Snow White project, because we can prolong the sleeping time of Snow White and take sufficient time for Prince to find Snow White. As a conclusion, the “Stop and Go” experiment was successful.
Ⅲ.mazEF System Assay ~Go & Stop~
In experimentⅡ, a toxin inhibits cell growth, and an antitoxin resuscitates it. However, what will happen when a toxin is expressed after the constitutive expression of antitoxin? Therefore, in the experiments of this section, we conducted a reciprocal experiment and named this experiment "Go & Stop".
We constructed
E. coli C: PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
E. coli A: Pcon - rbs - gfp (pSB6A1), Plac - rbs (pSB3K3)
E. coli G: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Pcon - rbs(weak) - mazE (pSB3K3)
E. coli F: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), Pcon - rbs - mazE (pSB3K3)
E. coli E: PBAD - rbs - mazF - tt - Pcon - rbs - gfp (pSB6A1), vector (pSB3K3).
result
Turbidity of E. coli G stopped earlier than that of E. coli F (Fig. 3), indicating that cellular mazE amount over MazF amount affects cytotoxity of MazF. In other words, the cytotoxity of MazF is determined stoichiometrically. Similarly, final RFU of E. coli G was lower than that of E. coli F. From the results of ExperimentⅠ and ExperimentⅡ, it is expected that mazEF can repeatedly control cell growth.
Calculation of the change of RFU of GFP / Turbidity per unit time (translation efficiency) indicates that the expression level of mazE correlated with the translation efficiency(Fig. 4).
Ⅳ.Control of Cell Growth
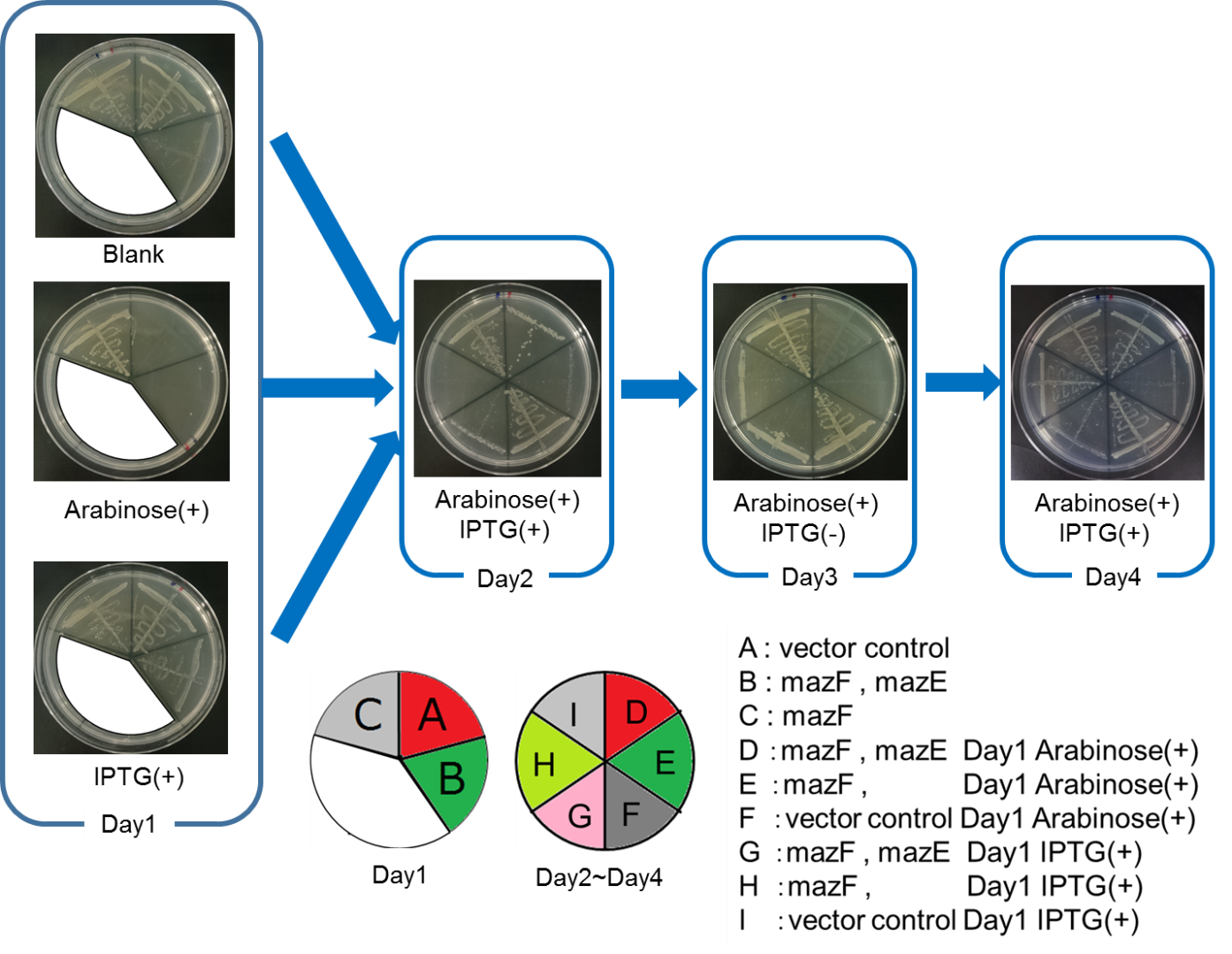
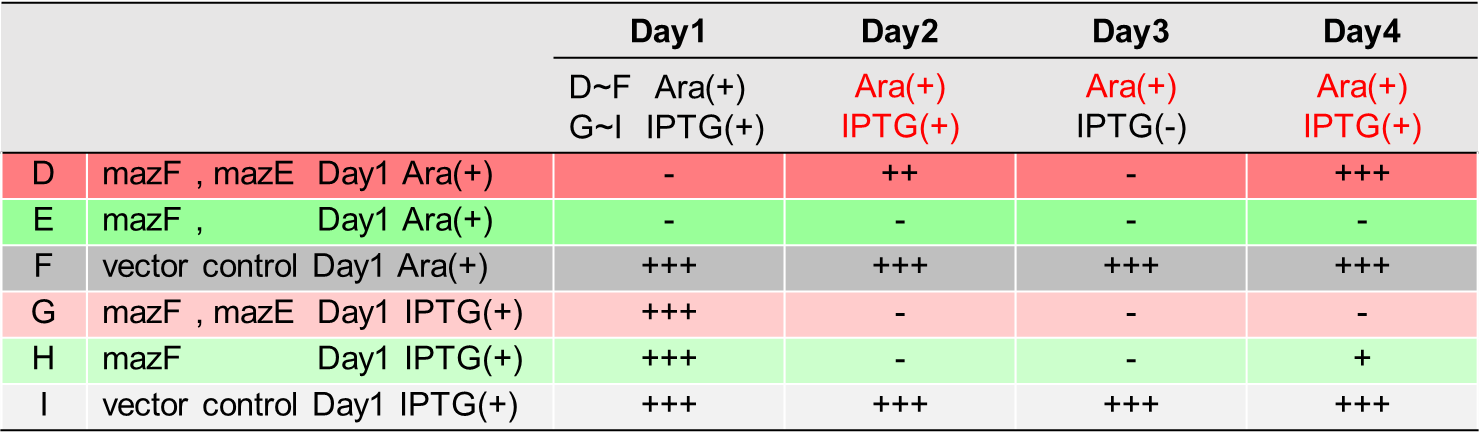
The control of cell growth by the mazEF system has been shown until the previous sections. In this section, we analyzed whether the “stop & go” experiment can be repeated many times.
We constructed
vector : PBAD - rbs (pSB6A1), Plac - rbs (pSB3K3)
MazF + MazE : PBAD - rbs - mazF (pSB6A1), Plac - rbs - mazE (pSB3K3)
MazF : PBAD - rbs - mazF(pSB6A1), Plac - rbs (pSB3K3).
result
From the result(Fig. 4., Fig. 5.), it was clarified that growth of E. coli cells was repeatedly controlled by expression of maze. The results of this experiment are very useful in our project, and it is expected to lead to new biotechnological applications.
reference
[1] Hazan, R., B. Sat, and H. Engelberg-Kulka. E. coli mazEF mediated cell death is triggered by various stressful conditions. J. Bacteriol. 2004 Dec;186(24):8295-8300.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1205
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961