Difference between revisions of "Part:BBa K1897024"
Phuongnguyen (Talk | contribs) |
Phuongnguyen (Talk | contribs) |
||
| Line 29: | Line 29: | ||
<p>Each individual components including the promoter, the coding sequence (ALR) and the terminator is isolated and amplified by PCR. After obtaining all the individual components, we ligated them by PCR overlap using appropriate primers (Figure 2). A second stop codon was added at the end of ALR sequence to enhance the efficiency of the translational termination.</p> | <p>Each individual components including the promoter, the coding sequence (ALR) and the terminator is isolated and amplified by PCR. After obtaining all the individual components, we ligated them by PCR overlap using appropriate primers (Figure 2). A second stop codon was added at the end of ALR sequence to enhance the efficiency of the translational termination.</p> | ||
[[File:62-33.PNG|thumb|center|800px|Figure 2: Construction of p62-33-ALR-Terminator by PCR overlap.<br> | [[File:62-33.PNG|thumb|center|800px|Figure 2: Construction of p62-33-ALR-Terminator by PCR overlap.<br> | ||
| − | + | The expected size, including Biobrick Prefix, Suffix and bases flanking restriction enzyme recognition sequences, is 1404 base pairs (bp). Lane 1 marks the DNA ladder. Lanes 2-5, which are replicates of the same reactions, show PCR products of p62-33-ALR-Terminator. | |
]] | ]] | ||
<h1>Characterization of RIOT sensor 8: p62-33-ALR-Terminator</h1> | <h1>Characterization of RIOT sensor 8: p62-33-ALR-Terminator</h1> | ||
| − | <p>RIOT sensors were transformed into E. coli Nissle ∆alr, a D-alanine auxotrophic mutant bacteria | + | <p>RIOT sensors were transformed into E. coli Nissle ∆alr, a D-alanine auxotrophic mutant bacteria [2]. In the absence of lactate, lldR binds to two operators in the promoter region and inhibit the expression of ALR. In the presence of lactate, lactate binds lldR, preventing its binding to the operators. Consequently, ALR, under the control of RIOT sensor promoter in the presence of L-lactate, will be expressed and rescue the transformed bacteria grown in media without D-alanine supplement (Figure 3). </p> |
[[File:Working mechanism.png|thumb|center|800px|Figure 3: Working mechanism of RIOT sensor. <br> | [[File:Working mechanism.png|thumb|center|800px|Figure 3: Working mechanism of RIOT sensor. <br> | ||
(A) In the absence of lactate, lldR binds to two operators in the promoter region and inhibit the expression of Alanine Racemase (ALR). <br> | (A) In the absence of lactate, lldR binds to two operators in the promoter region and inhibit the expression of Alanine Racemase (ALR). <br> | ||
(B) In the presence of lactate, lactate binds lldR, preventing its binding to the operators. Consequently, ALR is expressed and converts L-alanine to D-alanine which helps bacteria to survive. | (B) In the presence of lactate, lactate binds lldR, preventing its binding to the operators. Consequently, ALR is expressed and converts L-alanine to D-alanine which helps bacteria to survive. | ||
]] | ]] | ||
| − | <p>Therefore, the performance of RIOT sensors were assessed by the growth rate of transformed bacteria over a range of lactate concentration in M9 minimal media via natural logarithm (LN) of OD600. We used linear regression analysis to fit the data of bacterial growth rate using four data points of LN (OD600) values from 90 to 180 min of each sensor. This is because the growth rate of each constructs appeared to deviate from each other at the 90-minute time point. We postulated that induction of ALR by lactate might take about 90 minutes. The slope of each fitted linear line represents the growth rate of bacteria (P-value < 0.01).</p> | + | <p>Therefore, the performance of RIOT sensors were assessed by the growth rate of transformed bacteria over a range of lactate concentration in M9 minimal media via natural logarithm (LN) of OD600. We used linear regression analysis to fit the data of bacterial growth rate using four data points of LN (OD600) values from 90 to 180 min of each sensor. This is because the growth rate of each constructs appeared to deviate from each other at the 90-minute time point ([[Part:BBa_K1897021]] and [[Part:BBa_K1897024]]). We postulated that induction of ALR by lactate might take about 90 minutes. The slope of each fitted linear line represents the growth rate of bacteria (P-value < 0.01).</p> |
| − | <p>Since naturally occurring amino acids have an L-configuration in mammals and D-alanine was also shown to be absent from human plasma protein [3] [4], bacteria transformed with a well-controlled sensor will not be able to grow in the absence of both D-alanine and lactate. The results shown in Figure 4 RIOT sensors 8, p62-33-ALR-Terminator ([[Part:BBa_K1897024]]) with weak RBS appeared to exhibit very minimal leakiness. The transformed bacteria were unable to grow in the absence or at low lactate concentration (10<sup>-4</sup> M). The addition of lactate at high concentration (10<sup>-3</sup> and 10<sup>-2</sup> M) rescued the transformed bacteria grown in media without D-alanine. According to Table | + | <p>Since naturally occurring amino acids have an L-configuration in mammals and D-alanine was also shown to be absent from human plasma protein [3] [4], bacteria transformed with a well-controlled sensor will not be able to grow in the absence of both D-alanine and lactate. The results shown in Figure 4 RIOT sensors 8, p62-33-ALR-Terminator ([[Part:BBa_K1897024]]) with weak RBS appeared to exhibit very minimal leakiness. The transformed bacteria were unable to grow in the absence or at low lactate concentration (10<sup>-4</sup> M). The addition of lactate at high concentration (10<sup>-3</sup> and 10<sup>-2</sup> M) rescued the transformed bacteria grown in media without D-alanine. According to Table 1, the growth rate of bacteria transformed with sensor 8 grown in lactate concentration of 10<sup>-4</sup> M was not significantly different from zero. However, the growth and doubling time of sensor 8 were almost similar when lactate concentration increased from 10<sup>-3</sup> to 10<sup>-2</sup>. Further tuning is required, such as cloning RIOT sensor 8 in a low-copy plasmid, may increase the sensitivity of this sensor to small changes in lactate concentration. </p> |
[[File:62-33 LN 1.jpg|thumb|center|800px|Figure 4: Sensitivity of RIOT Sensor 8: p62-33-ALR-Terminator to lactate<br> | [[File:62-33 LN 1.jpg|thumb|center|800px|Figure 4: Sensitivity of RIOT Sensor 8: p62-33-ALR-Terminator to lactate<br> | ||
| − | D-alanine auxotrophic mutant bacteria were transformed with plasmids containing ALR sequence under the control of RIOT sensor 8: p62-33-ALR-Terminator ([[Part:BBa_K1897024]]). The overnight culture were spun down and resuspended with M9 minimal media without D-alanine supplement. Then, they were subjected to different treatments: without and with addition of D-alanine supplement (50 μg/ml) and with addition of lactate of 10-4 to 10-2 M. OD600 readings were measured for 6 hours. LN | + | D-alanine auxotrophic mutant bacteria were transformed with plasmids containing ALR sequence under the control of RIOT sensor 8: p62-33-ALR-Terminator ([[Part:BBa_K1897024]]). The overnight culture were spun down and resuspended with M9 minimal media without D-alanine supplement. Then, they were subjected to different treatments: without and with addition of D-alanine supplement (50 μg/ml) and with addition of lactate of 10<sup>-4</sup> to 10<sup>-2</sup> M. OD600 readings were measured for 6 hours. Mean LN(OD600) was calculated with N=3 and the error bars stand for SEM. |
]] | ]] | ||
<b>Table 1: Bacterial growth rate of RIOT sensor 8: p62-33-ALR-Terminator ([[Part:BBa_K1897024]])</b> | <b>Table 1: Bacterial growth rate of RIOT sensor 8: p62-33-ALR-Terminator ([[Part:BBa_K1897024]])</b> | ||
| − | <p>N = | + | <p>N = 3 ± SEM<br> |
Doubling time = LN (2) / Growth rate. | Doubling time = LN (2) / Growth rate. | ||
</p> | </p> | ||
Latest revision as of 18:24, 19 October 2016
p62-33-ALR-Terminator
The construct contains a shorter version of the promoter region for wild-type lldPRD operon (Part:BBa_K1897037) derived from (Part:BBa_K822000) with a weaker RBS flanked by non-coding sequences (Part:BBa_K1897031), followed by a coding sequence of Alanine Racemase (Part:BBa_K1172901) and a lambda t0 terminator (Part:BBa_K1897030).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 499
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 423
Illegal BamHI site found at 1125 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 541
Illegal AgeI site found at 841 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 298
Usage and Biology
This is RIOT sensor 8 which belongs to the RIOT sensor collection (Part:BBa_K1897019, Part:BBa_K1897020, Part:BBa_K1897021, Part:BBa_K1897022, Part:BBa_K1897023, Part:BBa_K1897024, Part:BBa_K1897025, Part:BBa_K1897026, Part:BBa_K1897027) and was designed to detect the increased level of L-lactate produced by tumor cells in the microenviroment. A lactate sensitive promoter obtained from 2015 ETH Zurich team was used as starting material. Our designed sensor has to be extremely sensitive to small changes in lactate concentration. This means that our sensor needs to have no basal expression and significant increase in reporter output (Alanine Racemase, ALR) in the presence of lactate. ALR is required for synthesis of D-alanine, an essential amino acid for bacterial survival [1].
We minimise the basal expression of our reporter by linking different strength RBS parts with:
- A shorter version of the promoter region for wild-type lldPRD operon (Part:BBa_K1897037, derived from Part Part:BBa_K822000) called “p62”, or
- The lldRO1-J23117-lldRO2 promoter (Part:BBa_K1847008) called “p70”, or
- The constitutively expressed promoter J23100 (Part:BBa_K314100).
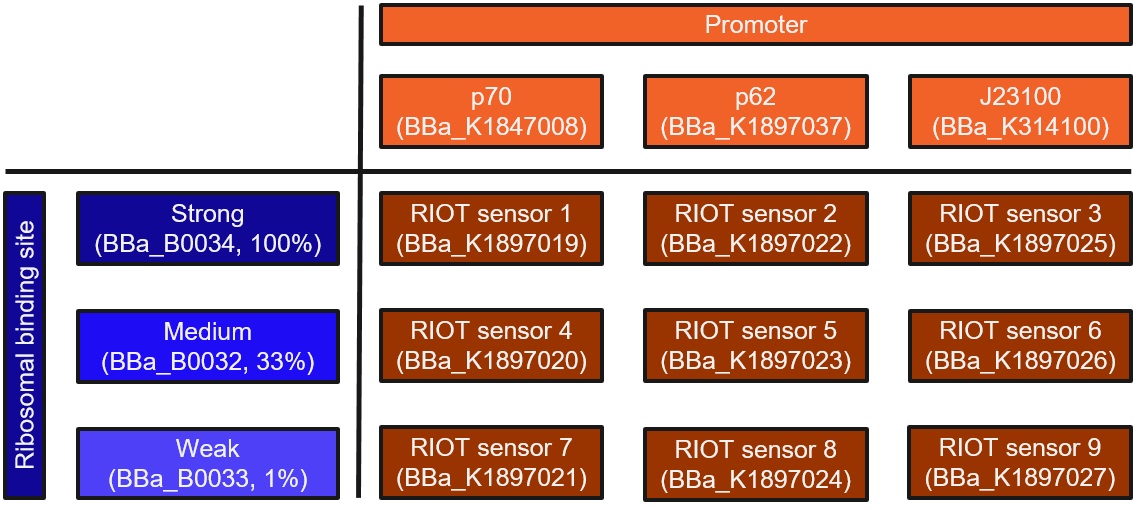
This collection is a combination of three promoters and three ribosomal binding sites (RBS) of different strengths. The promoters consists of a modified lactate sensor p70 (Part:BBa_K1847008), a shorter version of the promoter region for wild-type lldPRD operon (Part:BBa_K1897037, derived from Part:BBa_K822000) called p62 and a constitutively expressed promoter J23100 (Part:BBa_K314100).
Construction of RIOT sensor 8: p62-33-ALR-Terminator
Each individual components including the promoter, the coding sequence (ALR) and the terminator is isolated and amplified by PCR. After obtaining all the individual components, we ligated them by PCR overlap using appropriate primers (Figure 2). A second stop codon was added at the end of ALR sequence to enhance the efficiency of the translational termination.

The expected size, including Biobrick Prefix, Suffix and bases flanking restriction enzyme recognition sequences, is 1404 base pairs (bp). Lane 1 marks the DNA ladder. Lanes 2-5, which are replicates of the same reactions, show PCR products of p62-33-ALR-Terminator.
Characterization of RIOT sensor 8: p62-33-ALR-Terminator
RIOT sensors were transformed into E. coli Nissle ∆alr, a D-alanine auxotrophic mutant bacteria [2]. In the absence of lactate, lldR binds to two operators in the promoter region and inhibit the expression of ALR. In the presence of lactate, lactate binds lldR, preventing its binding to the operators. Consequently, ALR, under the control of RIOT sensor promoter in the presence of L-lactate, will be expressed and rescue the transformed bacteria grown in media without D-alanine supplement (Figure 3).
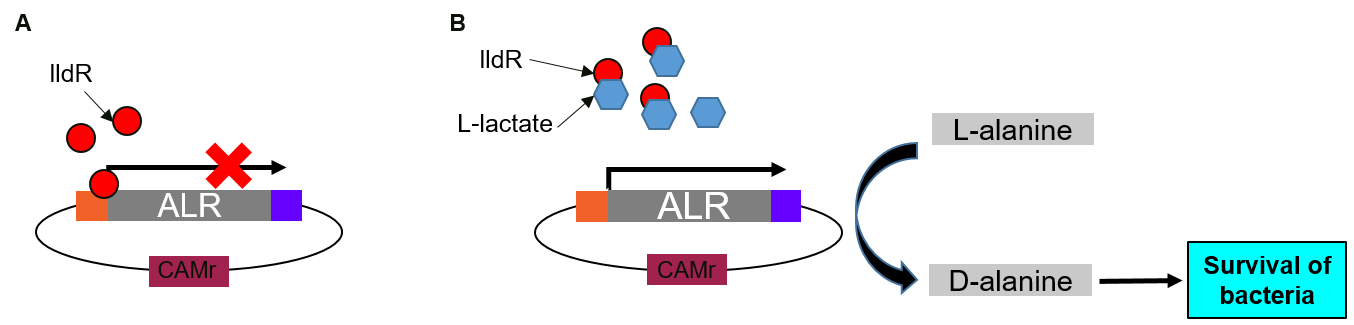
(A) In the absence of lactate, lldR binds to two operators in the promoter region and inhibit the expression of Alanine Racemase (ALR).
(B) In the presence of lactate, lactate binds lldR, preventing its binding to the operators. Consequently, ALR is expressed and converts L-alanine to D-alanine which helps bacteria to survive.
Therefore, the performance of RIOT sensors were assessed by the growth rate of transformed bacteria over a range of lactate concentration in M9 minimal media via natural logarithm (LN) of OD600. We used linear regression analysis to fit the data of bacterial growth rate using four data points of LN (OD600) values from 90 to 180 min of each sensor. This is because the growth rate of each constructs appeared to deviate from each other at the 90-minute time point (Part:BBa_K1897021 and Part:BBa_K1897024). We postulated that induction of ALR by lactate might take about 90 minutes. The slope of each fitted linear line represents the growth rate of bacteria (P-value < 0.01).
Since naturally occurring amino acids have an L-configuration in mammals and D-alanine was also shown to be absent from human plasma protein [3] [4], bacteria transformed with a well-controlled sensor will not be able to grow in the absence of both D-alanine and lactate. The results shown in Figure 4 RIOT sensors 8, p62-33-ALR-Terminator (Part:BBa_K1897024) with weak RBS appeared to exhibit very minimal leakiness. The transformed bacteria were unable to grow in the absence or at low lactate concentration (10-4 M). The addition of lactate at high concentration (10-3 and 10-2 M) rescued the transformed bacteria grown in media without D-alanine. According to Table 1, the growth rate of bacteria transformed with sensor 8 grown in lactate concentration of 10-4 M was not significantly different from zero. However, the growth and doubling time of sensor 8 were almost similar when lactate concentration increased from 10-3 to 10-2. Further tuning is required, such as cloning RIOT sensor 8 in a low-copy plasmid, may increase the sensitivity of this sensor to small changes in lactate concentration.

D-alanine auxotrophic mutant bacteria were transformed with plasmids containing ALR sequence under the control of RIOT sensor 8: p62-33-ALR-Terminator (Part:BBa_K1897024). The overnight culture were spun down and resuspended with M9 minimal media without D-alanine supplement. Then, they were subjected to different treatments: without and with addition of D-alanine supplement (50 μg/ml) and with addition of lactate of 10-4 to 10-2 M. OD600 readings were measured for 6 hours. Mean LN(OD600) was calculated with N=3 and the error bars stand for SEM.
Table 1: Bacterial growth rate of RIOT sensor 8: p62-33-ALR-Terminator (Part:BBa_K1897024)
N = 3 ± SEM
Doubling time = LN (2) / Growth rate.
| Lactate concentration (M) | Growth rate (min-1) | Doubling time (min) | R square |
|---|---|---|---|
| 10-4 | No growth was observed | ||
| 10-3 | 0.00821 ± 0.000490 | 84.7 ± 5.06 | 0.989 |
| 10-2 | 0.00830 ± 0.000562 | 83.9 ± 5.68 | 0.986 |
References
- Walsh, C. T. (1989). Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. Journal of biological chemistry, 264(5), 2393-2396.
- In Young Hwang, Elvin Koh, Adison Wong, John C. March, William E. Bentley, Yung Seng Lee and Matthew Wook Chang (2016). Engineered probiotic microbes eliminate and prevent pathogen infection in the mammalian gut. Manuscript submitted.
- Nagata, Y., Masui, R., & Akino, T. (1992). The presence of free D-serine, D-alanine and D-proline in human plasma. Experientia, 48(10), 986-988.
- Hoeprich, P. D. (1965). Alanine: Cyeloserine Antagonism. VI. Demonstration of D-Alanine in the Serum of Guinea Pigs and Mice. Journal of Biological Chemistry, 240, 1654-60.
