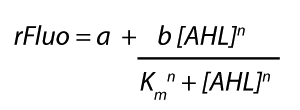Difference between revisions of "Part:BBa C0062:Experience"
(→First-order crosstalk) |
(→First-order crosstalk) |
||
| Line 40: | Line 40: | ||
== First-order crosstalk == | == First-order crosstalk == | ||
| − | + | In the first order crosstalk section we describe activation of [https://parts.igem.org/Part:BBa_R0062 pLux] due to [https://parts.igem.org/Part:BBa_C0062 LuxR] binding to inducers different from [[3OC6HSL|3OC6-HSL]] or [https://parts.igem.org/Part:BBa_R0062 pLux] itself binding a regulator-inducer pair different from [https://parts.igem.org/Part:BBa_C0062 LuxR]-[[3OC6HSL|3OC6-HSL]]. | |
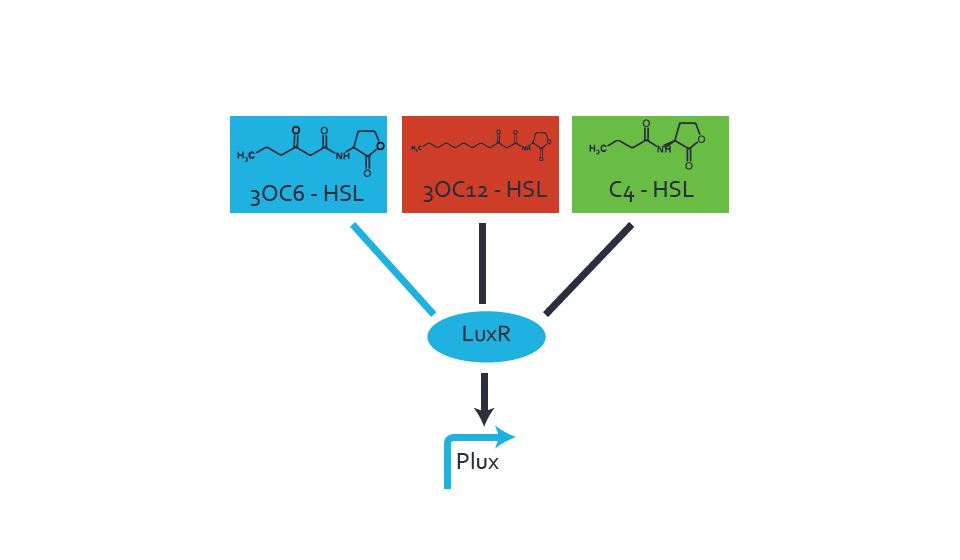
=== First Level crosstalk: LuxR binds to different HSL and activates the promoter Plux === | === First Level crosstalk: LuxR binds to different HSL and activates the promoter Plux === | ||
Revision as of 15:03, 24 October 2014
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_C0062
User Reviews
UNIQ800aa2db79897ae7-partinfo-00000000-QINU
|
••••
ETH Zurich 2014 |
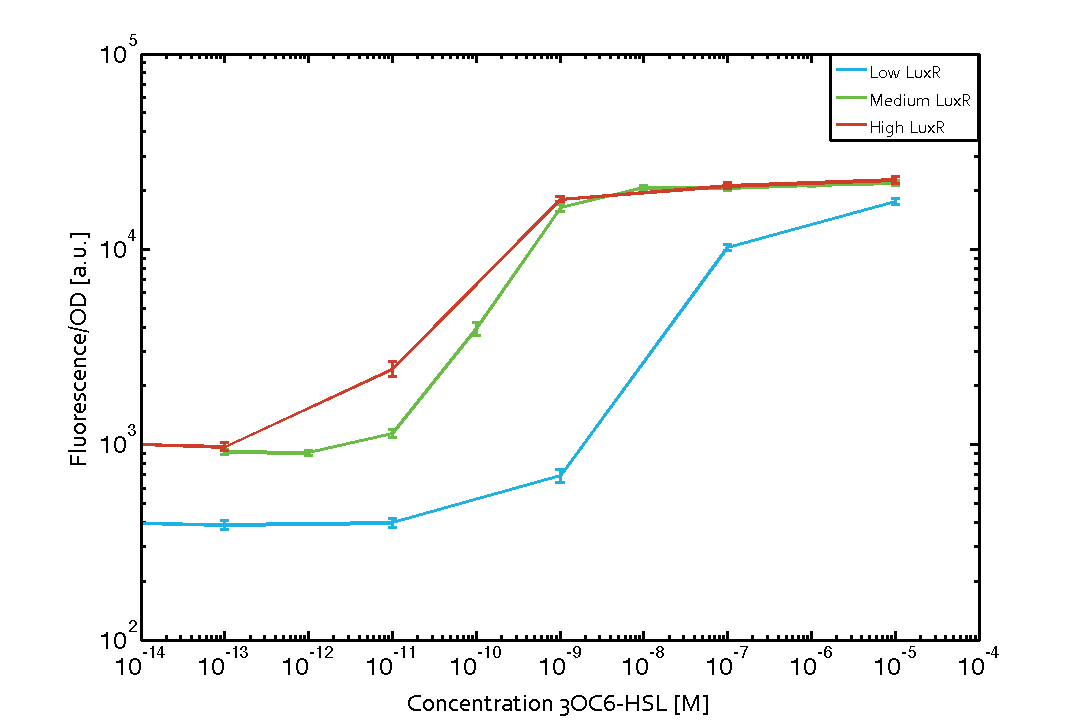
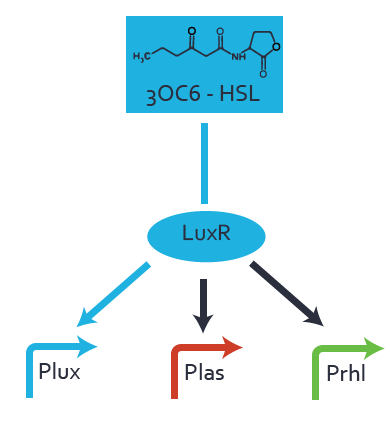
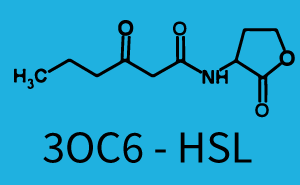
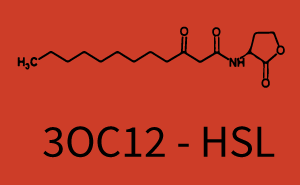
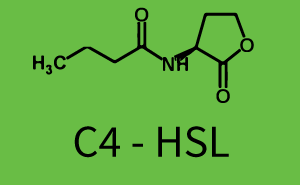
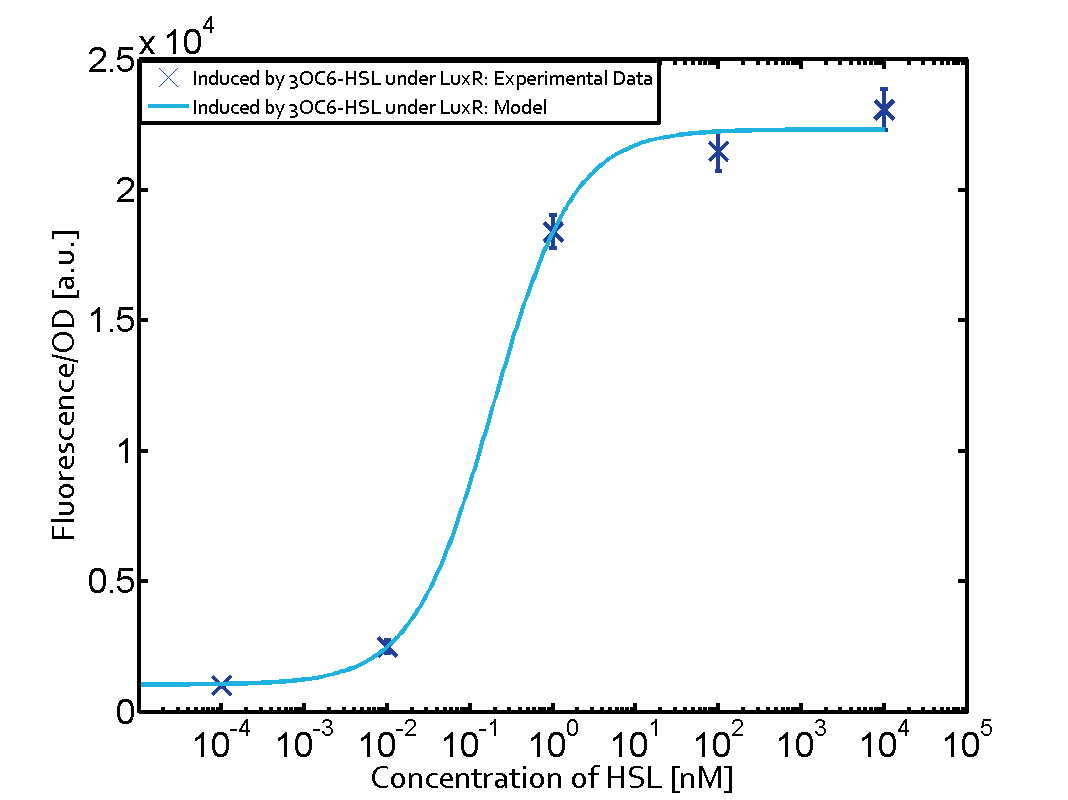
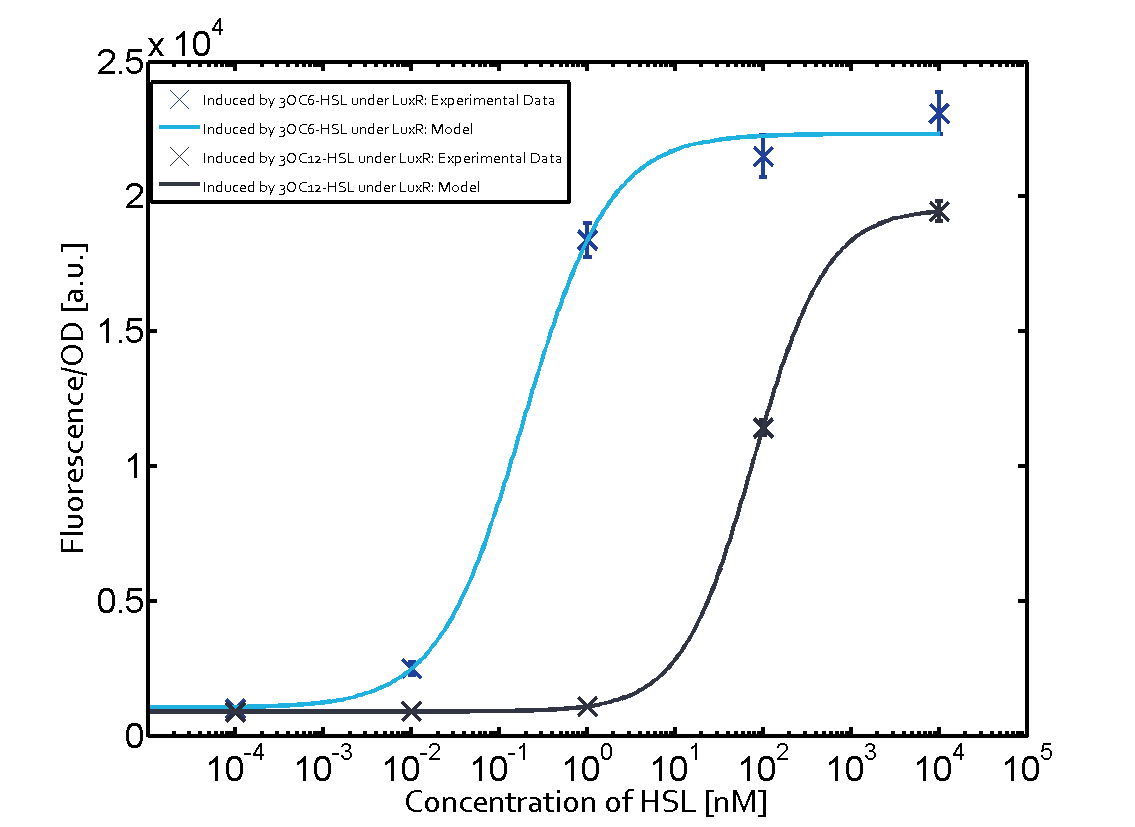
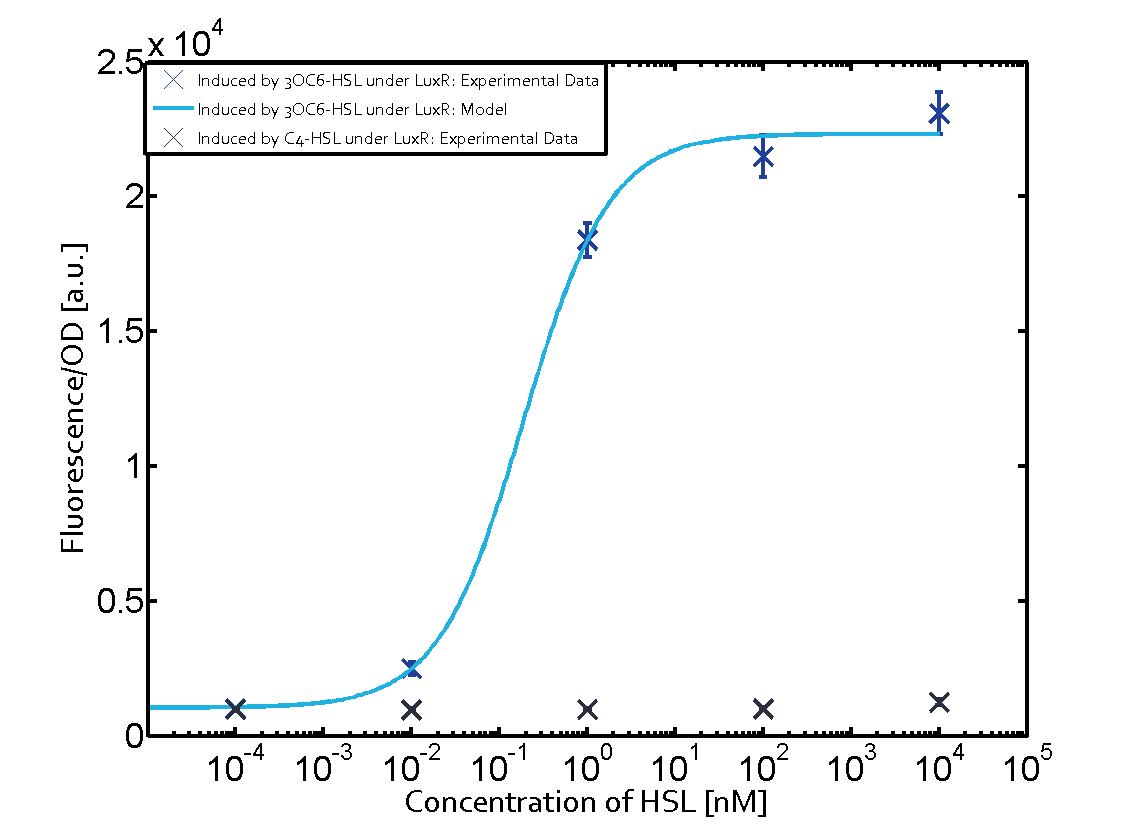
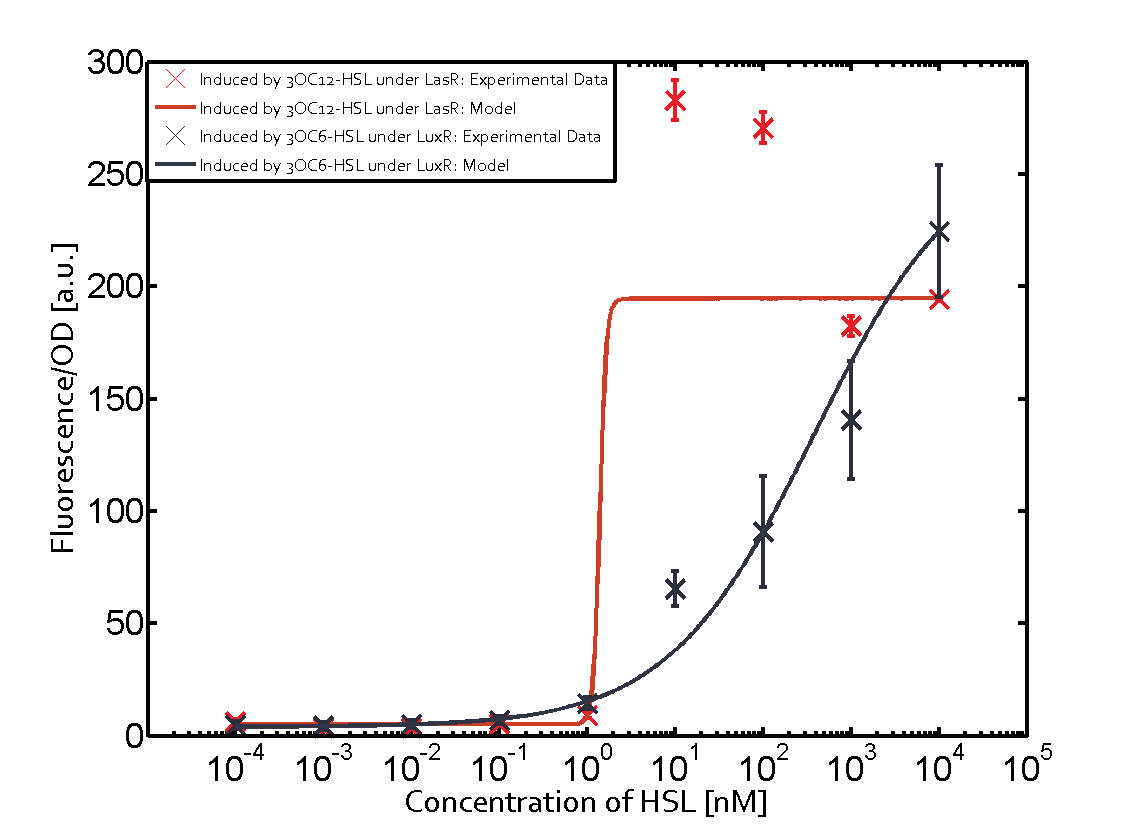
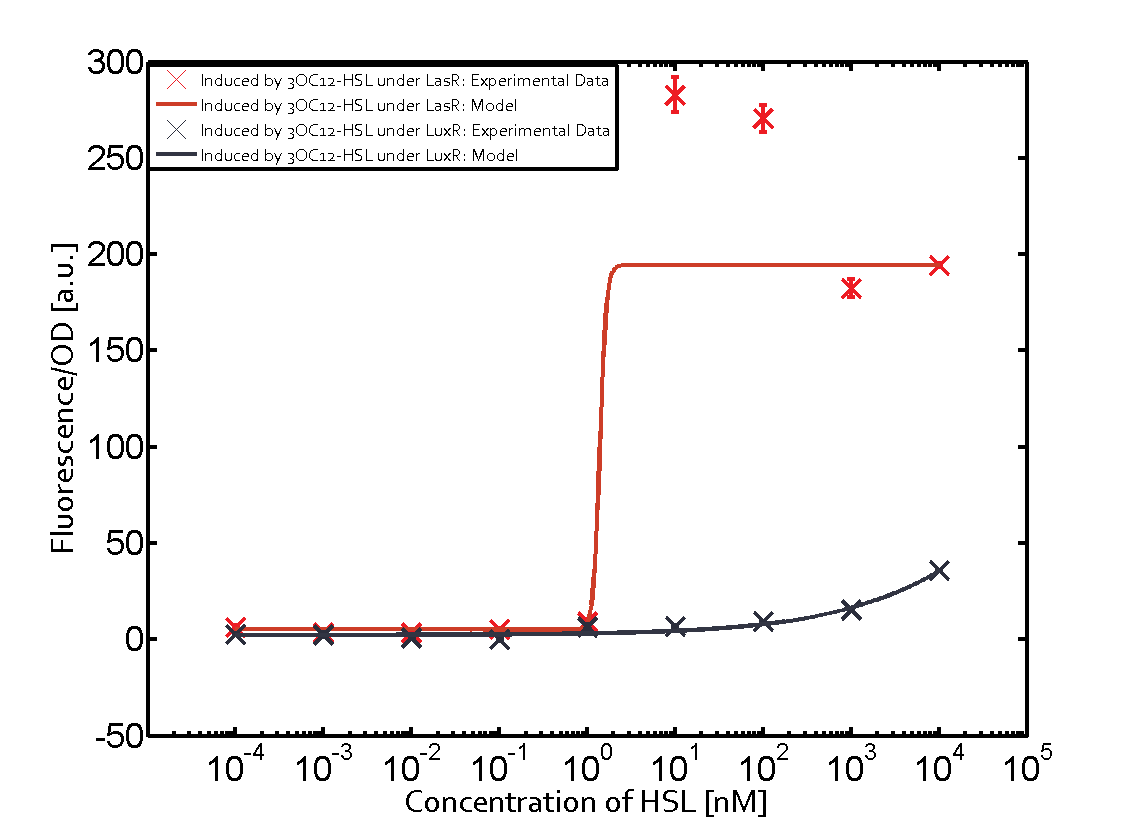
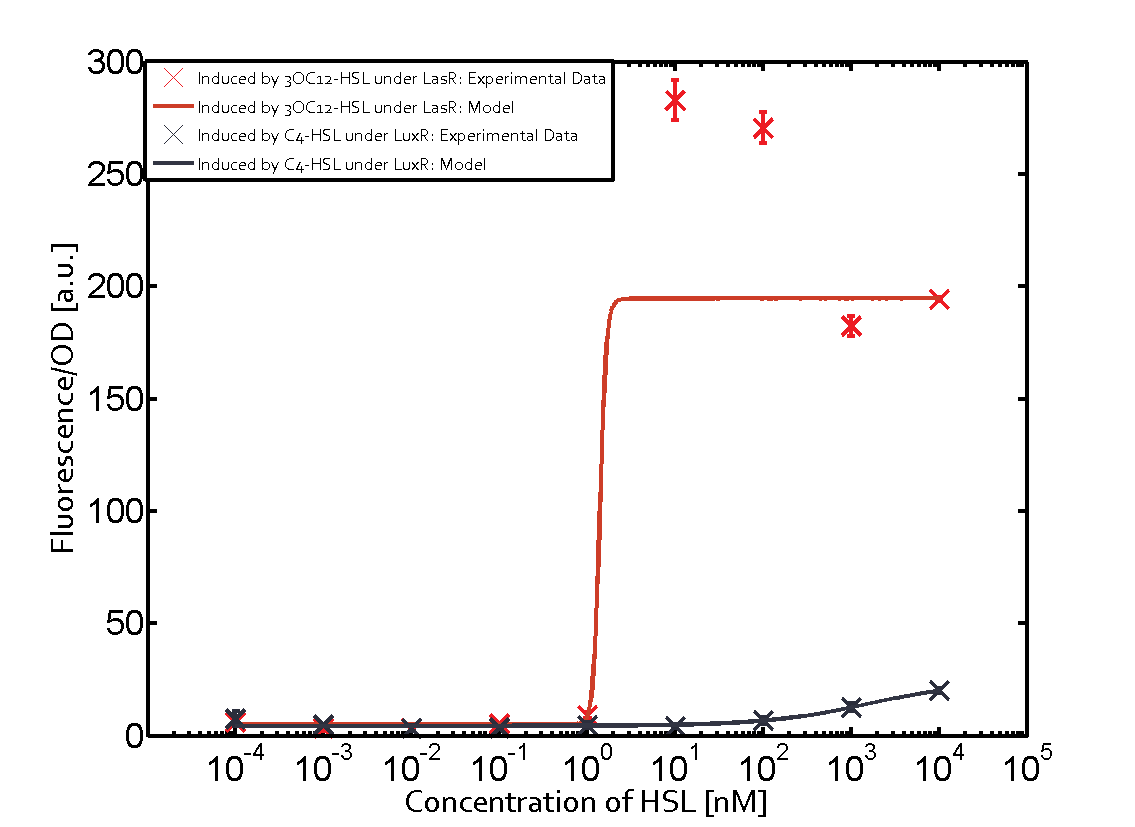
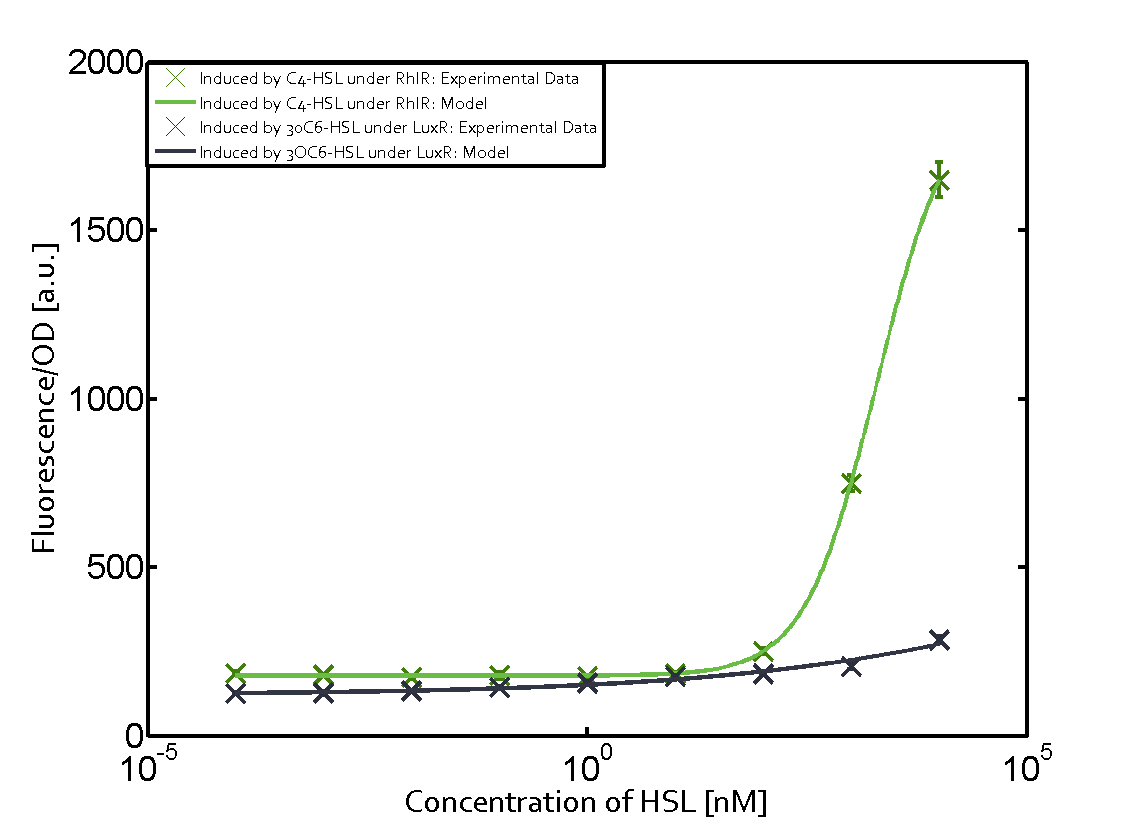
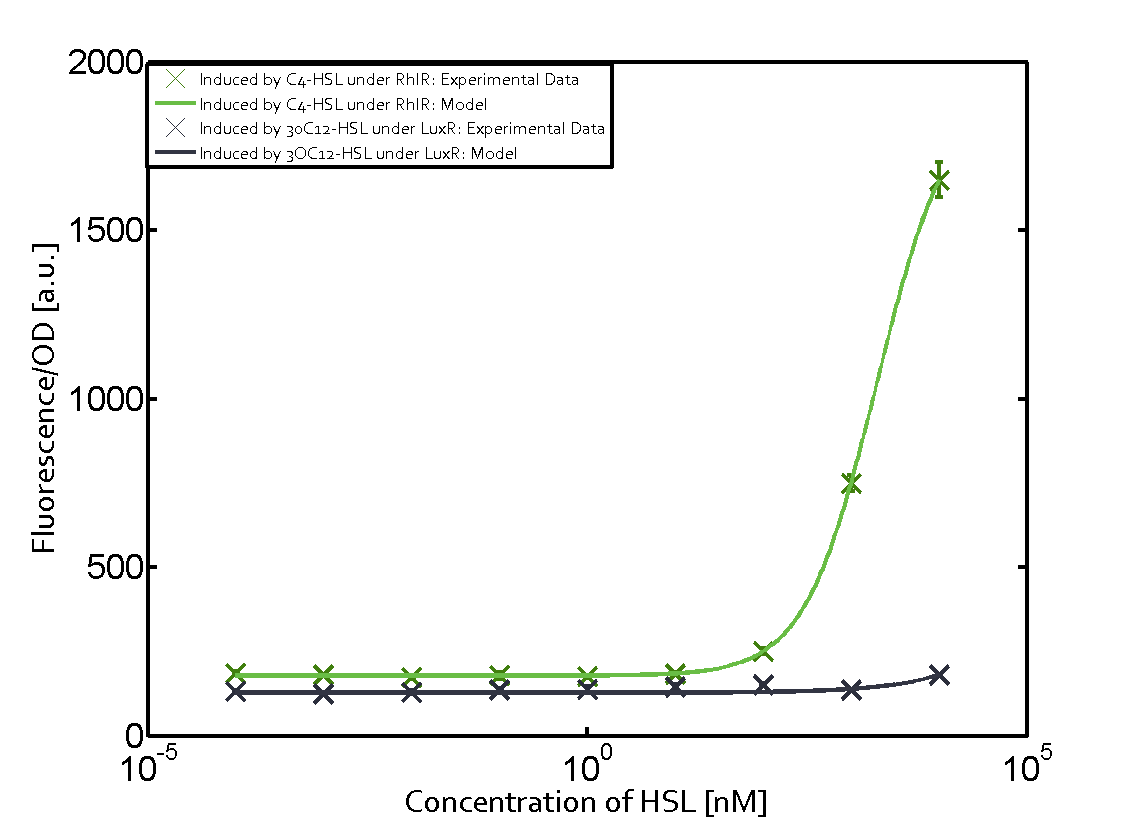
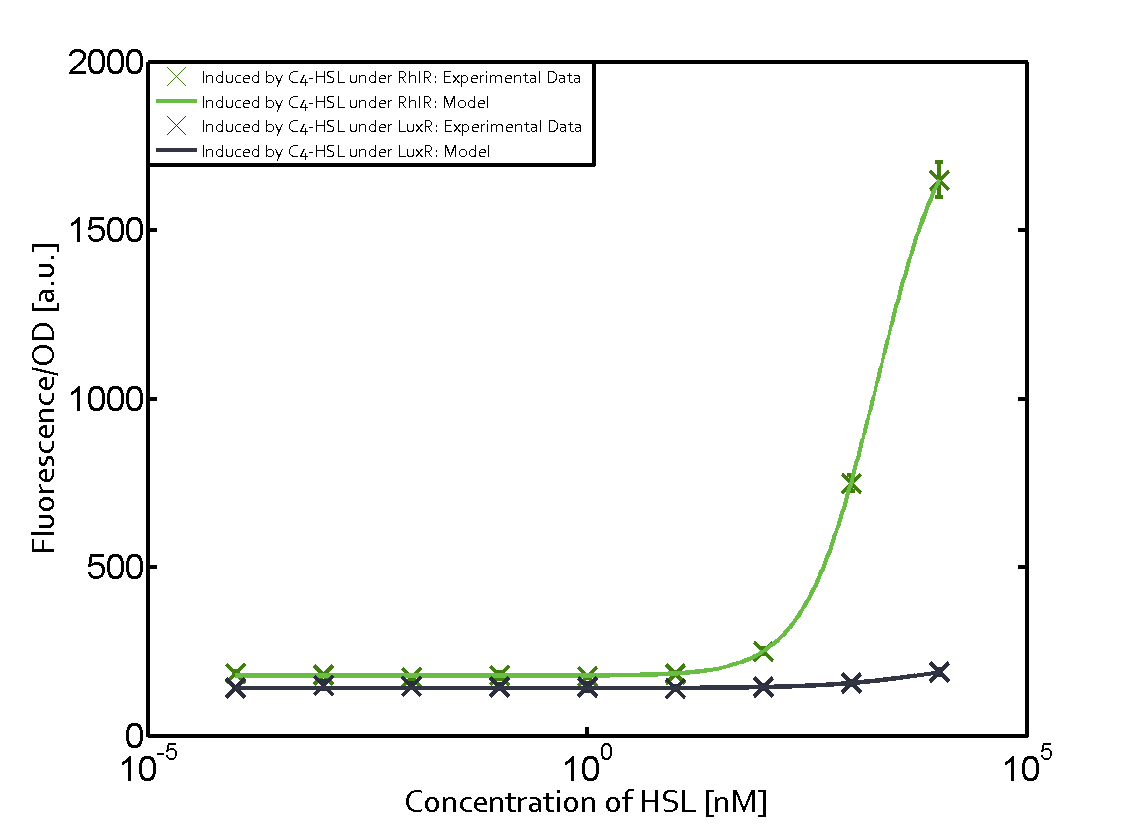
Characterization of the promoter's sensitivity to 3OC6-HSL depending on LuxR concentrationBackground informationResultsCharacterization of two-order crosstalkBackground informationHere, we focus on the characterization of crosstalk of LuxR with different HSLs and further crosstalk of LuxR-3OC6-HSL with the three promoters - pLux, pLas, and pRhl. In the following, we describe all the different levels of crosstalk we have assessed. First-order crosstalkIn the first order crosstalk section we describe activation of pLux due to LuxR binding to inducers different from 3OC6-HSL or pLux itself binding a regulator-inducer pair different from LuxR-3OC6-HSL. First Level crosstalk: LuxR binds to different HSL and activates the promoter PluxIn the conventional system 3OC6-HSL binds to its corresponding regulator, LuxR, and activates the pLux promoter (figure 2, light blue). However, LuxR can potentially also bind to other AHLs and then activate pLux (figure 2, 3OC12-HSL in red and C4-HSL in green). Second Level crosstalk: LuxR binds to 3OC6-HSL, its natural HSL, and activates different promotersSecond order crosstalk: Combination of both cross-talk levelsOther regulatory proteins, like LuxR, bind to different HSL and activates the promoter. Results
Modeling crosstalkEach experimental data set was fitted to an Hill function using the Least Absolute Residual method. The fitting of the graphs was performed using the following equation :
| ||||||||||||||||||||||||||||||||||||
|
•••••
SUN(Tsinghua) |
Part was sequenced and functional. LuxR was used in our Portable Pathogen Detector. |
|
•••••
wmholtz |
Using this part, I have successfully constructed and tested a quorum sensing circuit in E. coli. |
|
•••••
Youri |
This part was used and tested as a subpart in K546000, K546001, K546002, K546003, K546005 and K546546. This part functioned in all cases. |
|
Kevin (iGEM Braunschweig 2013) |
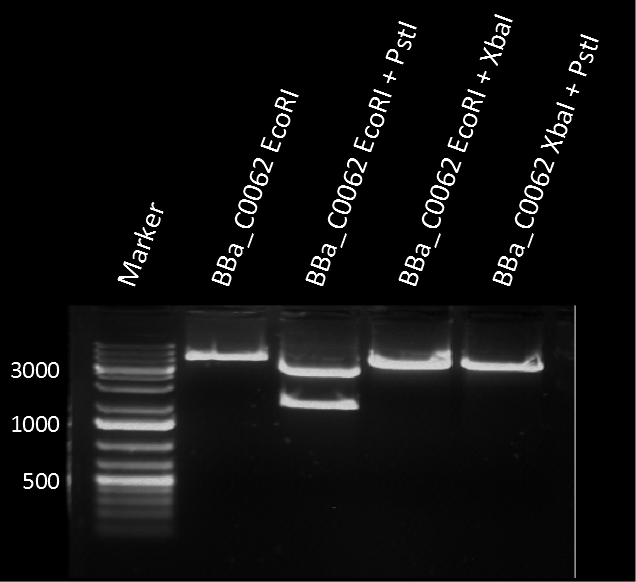
The plasmid pSB1C3 BBa_C0062 from the 2013 distribution Kit was transformed in E. coli XL1 BlueMRF. Sequencing with standard verification primer VF2 confirmed matching sequence of backbone DNA up to the EcoRI restriction site. The rest of the sequence (not shown) does not match the registry entry.
96 145
pSB1C3 LuxR (96) GAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATT
C0062 VF2 (1) GAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATT
146 195
pSB1C3 LuxR (146) TCTGGAATTCGCGGCCGCTTCTAGAGATGAAAAACATAAATGCCGACGAC
C0062 VF2 (51) TCTGGAATTCGACGCAA-TGGGTGCGCTGTCTACTAAATACAACGACACC
196 245
pSB1C3 LuxR (196) ACATACAGAATAATTAATAAAATTAAAGCTTGTAGAAGCAATAATGATAT
C0062 VF2 (100) CCGGAAAAAGCCTCCCGTACTTACGACGCTCACCGTGACGGTTTCGTTAT
246
pSB1C3 LuxR (246) TAATCAATGC...
C0062 VF2 (150) CGCTGGCGGC...
A restriction assay (Figure 1) showed that the sequenced part has no XbaI restriction site following the EcoRI site indicating another part in front of BBa_C0062 with a length of at least 1000 bp. |
UNIQ800aa2db79897ae7-partinfo-00000007-QINU