Part:BBa_K3425009
pSB4K01: pOdd1 Loop Vector based on pSB4K5
This part, pSB4K01, is a low copy number pOdd1 Loop vector based on pSB4K5.
New pOdd backbones for Level 3 assemblies were constructed using the Gibson assembly method. These are three sets of pOdd plasmids which suit different situations:
- pSB3K0# (BBa_K3425005 to BBa_K3425008): medium copy number (10-12 copies)
- pSB4K0# (BBa_K3425009 to BBa_K3425012): low copy number (~5 copies)
- pSB4A0# (BBa_K3425013 to BBa_K3425016): low copy number (~5 copies) with different resistance for cotransformations
The low copy number set of backbones with the selection marker for ampicillin resistance was specifically designed to allow double transformation of high (kanamycin) and low (ampicillin) copy number plasmid. It was also designed to allow triple transformation. Triple transformation of high (kanamycin), medium (chloramphenicol) and low (ampicillin) copy number plasmid can be useful in the systems where the extreme differences in the levels of the components are needed.
Low copy number plasmids (~5) are useful when the medium copy number plasmid (10-12) is too high. These plasmids can be used, for example, for really large constructs or as an alternative to genomic integration (in terms of copy number).
Medium copy number pOdd Loop Vectors (for Level 3)
When Level 2 multi-transcriptional units (MTUs) are not enough to hold your whole system, you might want to construct higher level assemblies. The current pOdd backbones, pSB1K0#, are high copy number backbones (100-300 copies) which are not suitable for Level 3 assemblies. The metabolic burden caused by the expression of multiple transcriptional units in a high copy number plasmid can be too overwhelming for the cell, which might make the plasmid unstable and prone to mutation or loss [1]. For this reason, new sets of medium (10-12 copies) and low (~5 copies) copy number pOdd backbones were designed and assembled.
This page details the set of low copy number backbones with Kanamycin resistance. This set keeps the alternating Cm-Kan resistances on Type IIS backbones. The pSB4K01-pSB4K04 series of plasmids is based on the pOdd set of iGEM Type IIS Vectors and are built upon the pSB4K5 plasmid backbone. When four Level 2 MTUs are assembled into these plasmid backbones (with BBa_J04454), the multi-MTU will be flanked by SapI restriction sites and the same 3 bp Fusion Sites as Level 1 assemblies.
| Part number | Name | Loop Name | Fusion Site 5' | Multi-MTU | Fusion Site 3' |
|---|---|---|---|---|---|
| BBa_K3425009 | pSB4K01 | pOdd1 | ATG | Multi-MTU 1 | GCA |
| BBa_K3425010 | pSB4K02 | pOdd2 | GCA | Multi-MTU 2 | TAC |
| BBa_K3425011 | pSB4K03 | pOdd3 | TAC | Multi-MTU 3 | CAG |
| BBa_K3425012 | pSB4K04 | pOdd4 | CAG | Multi-MTU 4 | GGT |
The iGEM Type IIS assembly standard is based on MoClo and Loop
- A Modular Cloning System for Standardized Assembly of Multigene Constructs
- Loop assembly: a simple and open system for recursive fabrication of DNA circuits
Note: This section was partially adapted from pSB1K01. For more information on Level 1 assemblies, you can visit pSB1K01. For more information as well as all our experience in this cloning method you can visit Team Uppsala 2020's iGEM Type IIS standard guidebook.
This part is part of iGEM Uppsala 2020's iGEM Type IIS standard collection.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3428
Illegal PstI site found at 12 - 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3428
Illegal PstI site found at 12 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3428 - 23INCOMPATIBLE WITH RFC[23]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3428
Illegal PstI site found at 12 - 25INCOMPATIBLE WITH RFC[25]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3428
Illegal PstI site found at 12 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal SapI site found at 3434
Illegal SapI.rc site found at 5
Gibson assembly of pOdd Loop vectors
Each set of new pOdd backbones comprises four plasmids which have the mRFP1 device for pOdd plasmids (BBa_J04454) flanked by the corresponding Type IIS fusion sites substituting the BioBrick sites of the base plasmid. This set, pSB4K0#, is derived from the pSB4K5 plasmid and has a very low copy number (~5 copies).
Gibson assembly was used to construct these backbones, and the details can be found in the Design page of each of these parts. Afterwards, it was needed to screen for plasmids which contained the mRFP1 device and had the correct antibiotic resistance and copy number. Antibiotic resistance was screened by plating the transformation, the presence of the mRFP1 device by PCR and the copy number by sequencing the ori and by controlled plasmid extraction.
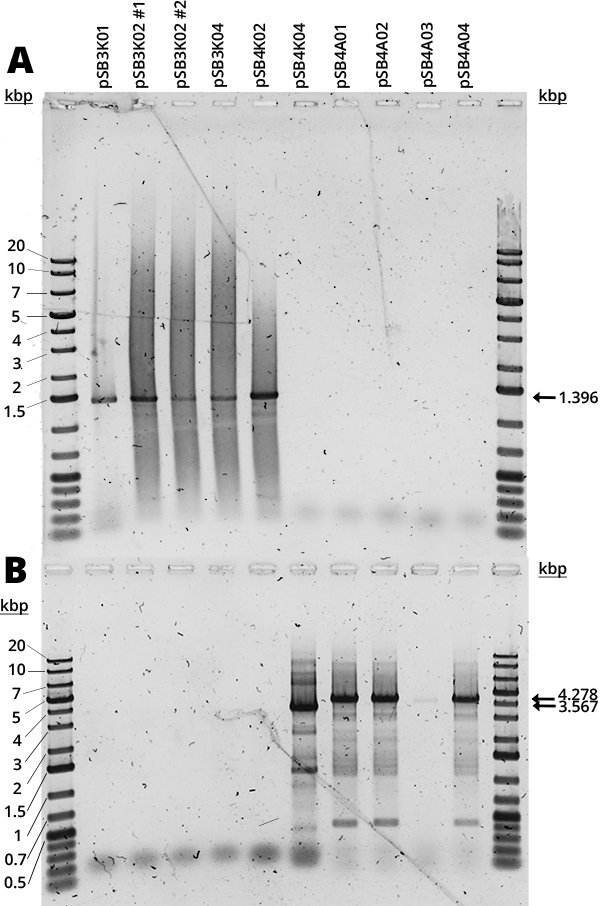
The Gibson assembly did not yield too many colonies and there was no time to repeat it, so there were no samples of pSB3K03, pSB4K01 and pSB4K02. In Figure 1 can be seen the amplification of the RFP device (A) and the full plasmid (B). The mRFP1 device (1396bp amplification) was present in all available pSB3K0# and pSB4K02. The full backbone amplification (3756bp for pSB3K0#, 3567bp for pSB4K0# and 4278bp for pSB4A0#) is positive for pSB4K04 and all pSB4A0#. Most samples (Figure 1, B) have multiple bands which we believe are non-specific and are due to the long extension times required for full backbone amplification, since the strongest band corresponds to the expected size.
The smear that can be seen in all samples (Figure 1) we suspect is due to the use of Diamond Dye (Promega), as it is something we have consistently observed with this DNA dye. Furthermore, it is concerning how in each gel half of the samples were negative, but it was different samples each time. Judging by the other results (Figures 1-4 and 6), it seems to be an error in the handling of samples. It would be good to repeat it, but there was no time. From the available results, however, it seems that all backbones are correct.
To evaluate the copy number of these new backbones, two different experiments were conducted. First, sequencing of the origin of replication and then controlled plasmid DNA extraction.
A region crucial for copy number regulation, the origin of replication (ori), was sequenced (Figures 2, 3 and 4).
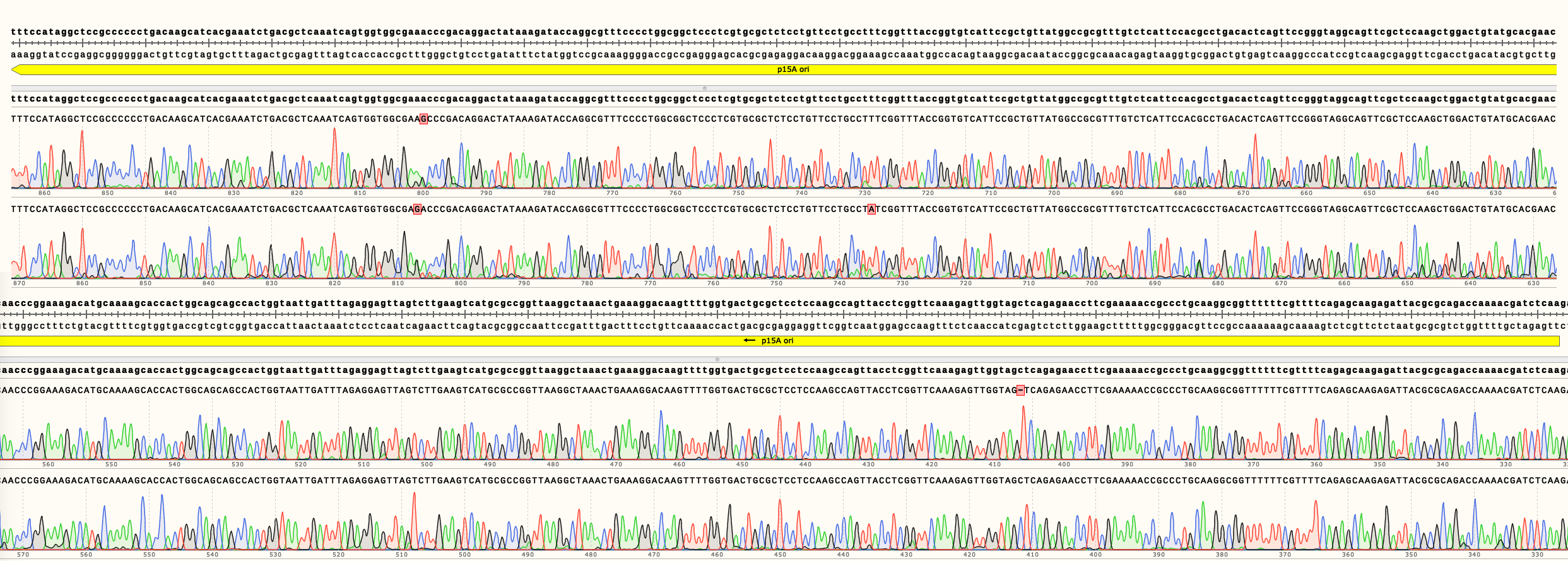
In Figure 4 can be seen the sequencing results of pSB3K02. It shows multiple single nucleotide changes in two different clones. It is unclear in which step the mutations were generated. Full length sequencing results are available in the Parts page.
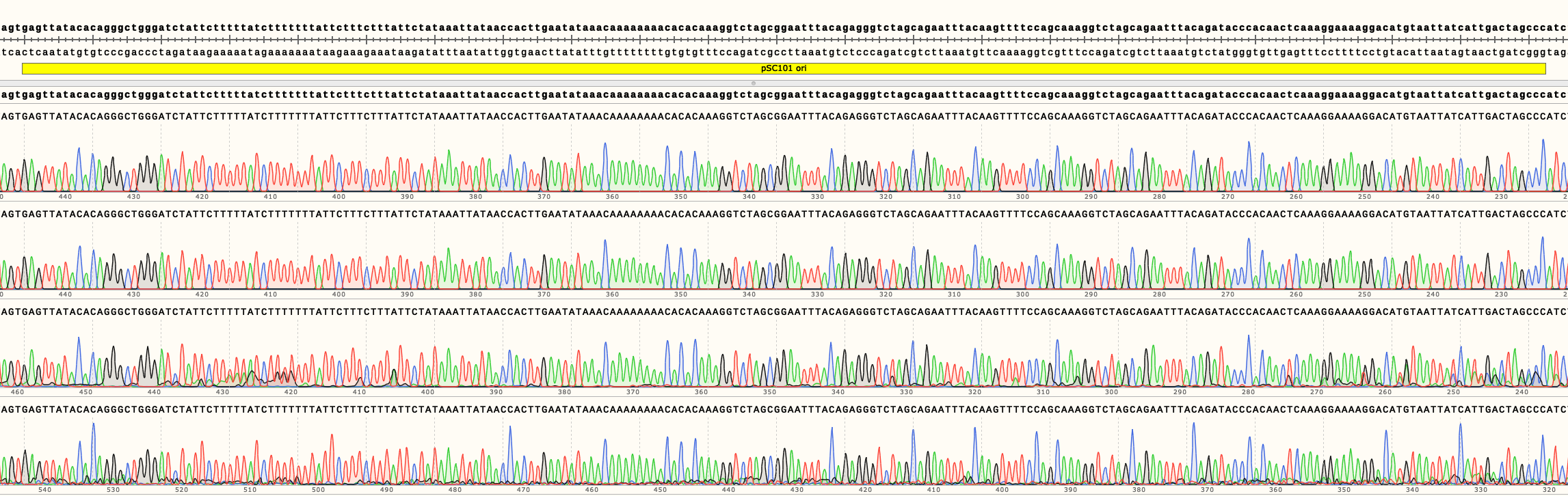
In Figure 3 can be seen the sequencing results of pSB4A01-4. They show no mutations in the ori of all four backbones. Full length sequencing results for all backbones are available in the Parts page.
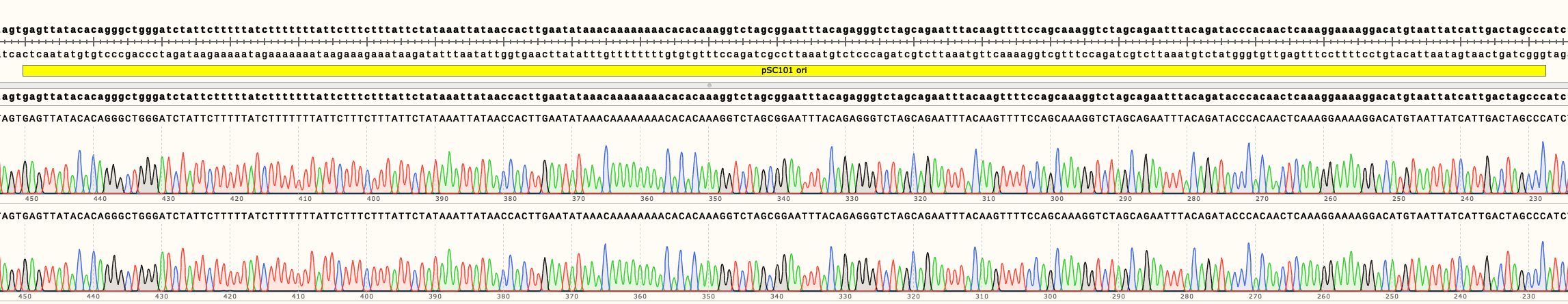
In Figure 4 can be seen the sequencing results of pSB4K02 and pSB4K04. They show no mutations in the ori. Full length sequencing results for both backbones are available in the Parts page.
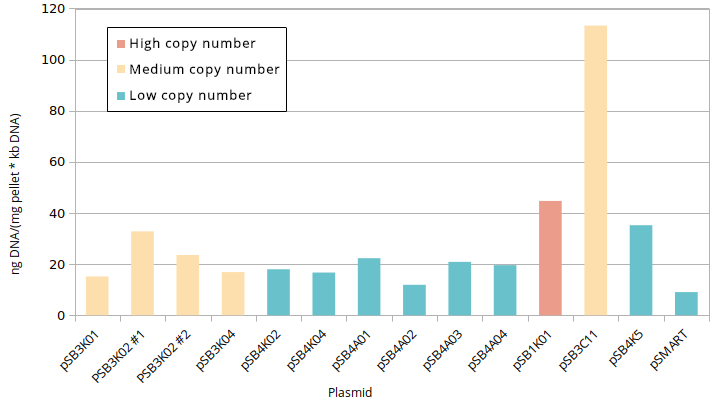
According to QIAGEN, a simple way to distinguish high and low copy number plasmids is to perform a miniprep [2]. Controlled plasmid extraction was conducted in order to see the difference between high (100-300), medium (15-20) and low (~5) copy number. This meant standardising plasmid DNA amounts to pellet weight that was used for its extraction, both the plasmids constructed by Gibson and controls for high (pSB1K01), medium (pSB3C11) and low (pSB4K5, pSMART [3]) copy number. Measuring the DNA was done in two different ways: via Nanodrop and via agarose gel electrophoresis.
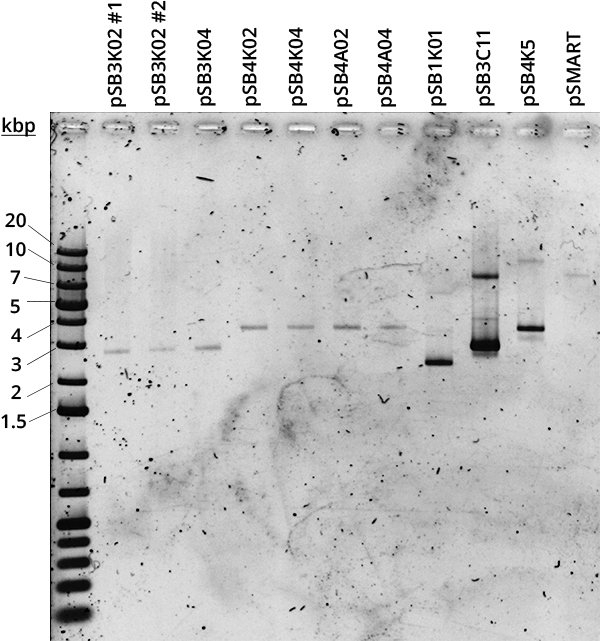
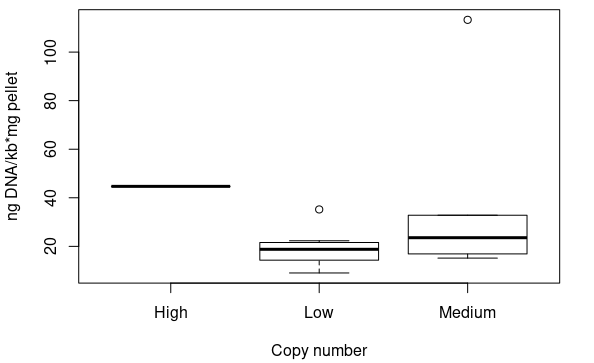
Standardised plasmid amounts were loaded on an agarose gel (Figure 5). Strangely, the controls (except pSMART) have much higher amounts of DNA than the plasmids constructed by Gibson, in spite of both groups having medium and low copy number plasmids. This is also seen in standardised Nanodrop results (Figure 6), which also shows that the control for medium copy number has a higher amount of DNA than the control for high copy number. A box plot of the results (Figure 7) suggests no difference between medium and low copy number.
An ANOVA test was conducted, however, to see if there was any difference between groups (high, medium, low copy number). The results were non-significant (p=0.52). Even with the clear medium copy number outlier removed (pSB3C11), the test was non-significant (p=0.21). Even if the sample size was small, there is far too much overlap in the standardised values of different copy numbers, and thus we can conclude that this method is not sensitive enough to distinguish between them.
This information can also be found in Team Uppsala 2020's Wet lab page. Sequencing results of this part with J04454 can be found in the Sequence Analysis section and the trace files are in the collection page as well as our Parts page.
References
[1] Jones, K. L., Kim, S.-W., and Keasling, J. D. (2000) Low-Copy Plasmids can Perform as Well as or Better Than High-Copy Plasmids for Metabolic Engineering of Bacteria. Metabolic Engineering. 2, 328â338
[2] How do I know if my plasmid is a high- or low copy number type? - QIAGEN [online] https://www.qiagen.com/no/resources/faq?id=1f42840e-fbd7-4734-b0cd-e17372a9e5a4&lang=en (Accessed October 24, 2020)
[3] CopyRight v2.0 BAC Cloning Kits (pSMART BAC and pEZ BAC) [online] https://www.lucigen.com/CopyRight-v2.0-BAC-Cloning-Kits-pSMART-BAC-and-pEZ-BAC/#subcat-tabs3 (Accessed October 21, 2020)
//plasmidbackbone/assembly/typeiis
| insert | RFP-device (J04455) |
| origin | pSC101 |
| resistance | K |
