Help:Standards/Assembly/Type IIS
- Registry Help Pages:
- TOC
- At-a-Glance
- FAQ
Accepted Standards: BioBrick RFC[10] | iGEM Type IIS RFC[1000]
Motivation
With a Type IIS assembly method, multiple parts can be assembled easily and reliably in a single reaction, and this can be scaled further to create more complex genetic circuits quickly, efficiently, and affordably.
A Type IIS assembly method uses Type IIS restriction enzymes, which are offsite cutters; they cut DNA at a specific distance from their recognition site. This allows for specified overhangs left by the Type IIS enzymes, which when intentionally designed can allow for multiple DNA fragments to be assembled together in the correct order and orientation.
Currently, there are several Type IIS assembly standards in use, many of these were designed for specific purposes. iGEM believes strongly in standards, and as we approach Type IIS assembly methods we want to support one that we believe will be useful, reliable, long-lasting and widespread for the synthetic biology community. The Type IIS assembly standard that iGEM accepts is based on MoClo and Loop.
The technical specifications of iGEM Type IIS (RFC1000) are laid out in the documentation below. This documentation will describe what makes a part compatible with the standard, and the requirements to create a sample of that part in Type IIS format. This is not meant to be a complete resource for Type IIS assembly, please see the previous work on MoClo and Loop for more information.
Overview
- Assembly Compatibility: Registry parts that adhere to iGEM Type IIS (RFC 1000) must not have Bsa1 and Sap1 Type IIS recognition sequences within their documented sequence. See Assembly Compatability below.
- Fusion Sites: RFC1000 specifies the fusion sites that will flank samples of basic parts (Level 0), transcriptional units (Level 1), and multi-transcriptional units (Level 2, 3). These are based on the MoClo/PhytoBricks and Loop Assembly standards.
The table below conforms iGEM's part abstraction (basic/composite) and part types, with the terminology (Levels 0, 1, 2, etc.) commonly used by other Type IIS assembly standards.
| Assembled into | ||||
|---|---|---|---|---|
| Level | Abstraction | Types | Loop Vector | Enzyme |
| Level 0 | Basic | Promoter, RBS, CDS, Terminator, etc. | pOdd (Level 1) | Bsa1 |
| Level 1 | Composite | Transcriptional units (devices, reporters, etc.) | pEven (Level 2) | Sap1 |
| Level 2 | Composite | Multi-transcriptional units (up to 4) | pOdd (Level 3) | Bsa1 |
| Level 3 | Composite | Multi-transcriptional units (up to 16) | pEven (Level 4) | Sap1 |
Technical Specifications
Assembly Compatibility
Parts that are compatible with iGEM Type IIS RCF1000 must not contain the Bsa1 and Sap1 Type IIS recognition sequences within their documented sequence. These two restriction enzymes are required for assembly. Parts that have these “illegal” recognition sites are not compatible with the standard.
| Enzyme | Type | Sequence (Forward) | Sequence (Reverse) |
|---|---|---|---|
| BsaI | Illegal | 5'...GGTCTC >>...3' 3'...CCAGAG >>...5' |
5'...<< GAGACC...3' 3'...<< CTCTGG...5' |
| SapI | Illegal | 5'...GCTCTTC >>...3' 3'...CGAGAAG >>...5' |
5'...<< GAAGAGC...3' 3'...<< CTTCTCG...5' |
| Enzyme | Sequence (Forward) | Sequence (Reverse) |
|---|---|---|
| BsaI | 5'...GGTCTC N NNNN...3' 3'...CCAGAG N NNNN...5' |
5'...NNNN N GAGACC...3' 3'...NNNN N CTCTGG...5' |
| SapI | 5'...GCTCTTC N NNN...3' 3'...CGAGAAG N NNN...5' |
5'...NNN N GAAGAGC...3' 3'...NNN N CTTCTCG...5' |
In order to check for Type IIS compatibility, view the Assembly Compatibility box below a part's sequence and features. RFC1000 is the draft name for iGEM's Type IIS standard.
See here for more general information on assembly compatibility.
Why were these restriction enzymes selected?
One benefit of Type IIS assembly standards is that the assembly method requires only two restriction enzymes for creation of basic parts (Level 0s), assembly of composite parts or transcriptional units (Level 1s), and multiple transcriptional units (Level 2+). Fewer illegal restriction sites means it will be easier to make compatible parts. Other Type IIS assembly standards may require different Type IIS restriction enzymes (AarI, Bsmb1, Bbs1, etc.), but in order to ensure ease of making parts compatible, the RFC1000 standard only designates Bsa1 and Sap1 as “illegal” restriction sites.
Why Bsa1?
Most, if not all, Type IIS assembly standards use Bsa1 for basic parts (Level 0) , so this ensures maximum compatibility.
Why Sap1?
In addition to Bsa1, Loop Assembly requires Sap1. While this restriction enzyme site only leaves a 3bp overhang, it has a 7bp recognition site, which theoretically will make it rarer, and make standardization of parts easier. For iGEM Type IIS, we have also modified the PhytoBricks Universal Acceptor plasmid, so that users can PCR their Level 0 parts into the iGEM Type IIS Universal Acceptor using Sap1. Cost analysis [link] has shown that Sap1, while initially more expensive than BioBrick enzymes, is within the price range of other Type IIS enzymes, and when scaled up to several rounds of multi-part assembly is affordable.
What about the BioBrick RFC10 restriction enzymes?
Your Type IIS RFC1000 parts do not need to be BioBrick RFC10 compatible. Your BioBrick parts do not need to be Type IIS RFC1000 compatible. However, if your part is compatible with both, iGEM and future users will be able to synthesize samples in both assembly formats.
Basic Parts (Level 0s)
Fusion Sites
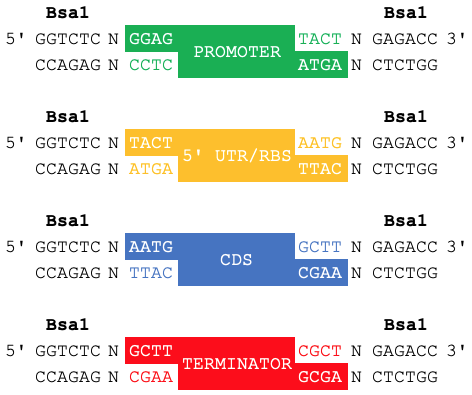
iGEM's Type IIS assembly standard specifies fusion sites that flank samples of the four basic parts types (Level 0).
| Fusion Site 5' | Part Type | Fusion Site 3' |
|---|---|---|
| GGAG | Promoter | TACT |
| TACT | 5'UTR | AATG |
| AATG | CDS | GCTT |
| GCTT | Terminator | CGCT |
Note, that these fusion sites do not belong on a basic part's documented sequence*. Instead, they belong to the prefix and suffix of the plasmid backbone that will be storing a sample of said part. There is an exception to this when documenting a CDS as your CDS will have a start codon (ATG). This ATG is also used in the 5' fusion site for all CDSs (AATG).
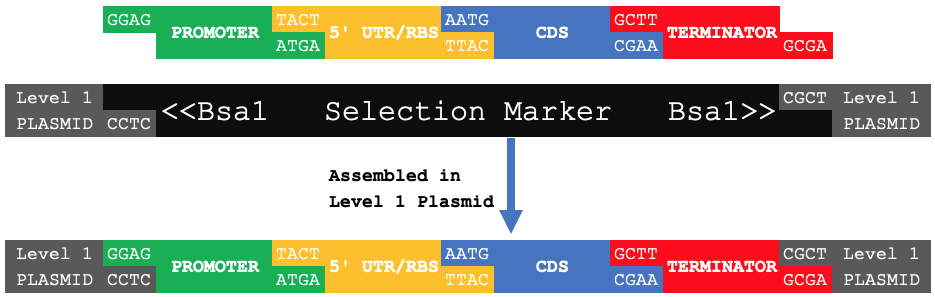
When samples of these basic parts are cut with Bsa1 and assembled together into a Level 1 plasmid backbone, it results in a composite part (Level 1), specifically a transcriptional unit, with the fusion sites now becoming scars within the composite part's sequence.
| Level 1 Assembly | ||||||||
|---|---|---|---|---|---|---|---|---|
| GGAG | Promoter | TACT | 5'UTR | AATG | CDS | GCTT | Terminator | CGCT |
| GGAG | Transcriptional Unit | CGCT | ||||||
Why were these fusion sites selected?
These fusion sites were outlined in the original MoClo paper (Weber, et al. 2011). They have since been adopted in other Type IIS assembly standards. As an example, these MoClo fusion sites completely overlap with the PhytoBricks standard. These fusion sites have also been tested against NEB’s 4bp ligation fidelity tool [link], finding them to be completely reliable when used as a set in a reaction.
To ensure maximum compatibility, and because of a long history of use, iGEM has adopted these fusion sites for four basic parts types (Promoter, RBS, CDS, Terminator).
iGEM has elected to not define additional fusion sites for other parts and sub-parts. These can be selected by the user depending on their assembly approach, or as laid out by other Type IIS methods like the PhytoBricks standard.
Can I use other fusion sites in my assembly?
Yes. Your lab or collection of parts may follow a different Type IIS assembly standard that specifies different fusion sites, or you may also be using assemblies with more than four parts and would need to use additional fusion sites.
Most importantly, though, your part's documet you can use your preferred fusion sites when assembling part samples. You When you're documenting your assembled composite part, make sure to enter in the exact scars you used for your assembly.
If you are submitting samples of your Type IIS parts to the Registry, then they must adhere to the iGEM Type IIS fusion sites. Note: Sample submission is no longer required. iGEM will syntheize samples of well-documented Type IIS parts according to the iGEM Type IIS standard.
Prefix and Suffix for Basic Parts
In a Level 0 plasmid backbone, a sample of these basic parts would be stored like so, with Bsa1 sites and fusion sites on the prefix and suffix flanking the part. You can create new basic parts through PCR and cloning into pSB1C00, the Universal Acceptor Vector for Level 0s.
| Prefix | Part Type | Suffix |
|---|---|---|
|
5' GGTCTC N GGAG CCAGAG N CCTC |
Promoter Promoter |
TACT N GAGACC 3' ATGA N CTCTGG |
|
5' GGTCTC N TACT CCAGAG N ATGA |
5'UTR 5'UTR |
AATG N GAGACC 3' TTAC N CTCTGG |
|
5' GGTCTC N AATG CCAGAG N TTAC |
CDS CDS |
GCTT N GAGACC 3' CGAA N CTCTGG |
|
5' GGTCTC N GCTT CCAGAG N CGAA |
Terminator Terminator |
CGCT N GAGACC 3' GCGA N CTCTGG |
When digested with Bsa1 and ligated with a Level 1 (pOdd) plasmid backbone in a one-pot reaction, these four basic parts will be assembled into a transcriptional unit (TU).
Transcriptional Units (Level 1s)
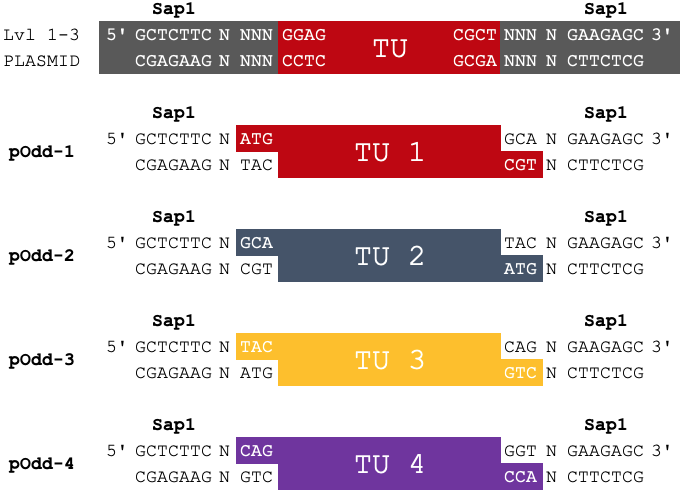
In Loop assembly, Level 0 basic parts can be assembled into a set of pOdd vectors. These Level 1 transriptional units will be flanked by Sap1 recognition sites and fusion sites, enabling further assembly of up to 4 transcriptional units.
Fusion Sites
| Fusion Site 5' | Transcriptional Unit (TU) | Fusion Site 3' |
|---|---|---|
| ATG | TU 1 | GCA |
| GCA | TU 2 | TAC |
| TAC | TU 3 | CAG |
| CAG | TU 4 | GGT |
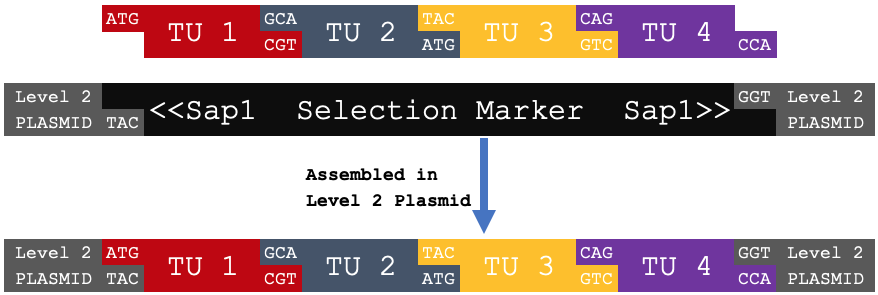
| Level 2 Assembly | ||||||||
|---|---|---|---|---|---|---|---|---|
| ATG | TU 1 | GCA | TU 2 | TAC | TU 3 | CAG | TU 4 | GGT |
| ATG | Multi-transcriptional Unit | GGT | ||||||
Prefix and Suffix
| Prefix | Transcriptional Unit (TU) | Suffix |
|---|---|---|
|
5' GCTCTTC N ATG CGAGAAG N TAC |
TU 1 TU 1 |
GCA N GAAGAGC 3' CGT N CTTCTCG |
|
5' GCTCTTC N GCA CGAGAAG N CGT |
TU 2 TU 2 |
TAC N GAAGAGC 3' ATG N CTTCTCG |
|
5' GCTCTTC N TAC CGAGAAG N ATG |
TU 3 TU 3 |
CAG N GAAGAGC 3' GTC N CTTCTCG |
|
5' GCTCTTC N CAG CGAGAAG N GTC |
TU 4 TU 4 |
GGT N GAAGAGC 3' CCA N CTTCTCG |
When digested with Sap1 and ligated into a Level 2 (pEven) plasmid backbone in a one-pot reaction, these four trancriptional units will be assembled into a multi-transcriptional unit (MTU).
References & Resources
Registry Plasmid Backbones
Universal Acceptor Vector (for Level 0)
- pSB1C00 with BBa_J04452
pOdd Loop Vectors (for Level 1)
Assembling into Level 1 Plasmds
pSB1K01-pSB1K04 series of plasmids are based on the pOdd set of Loop Vectors and are built upon the pSB1K3 plasmid backbone.
Note:
- Since pSB1K3 is a high-copy plasmid backbone, these vectors should not be used for Level 3 assemblies.
- Currently, Registry samples of these plasmid backbones have not been fully sequenced. Only their prefix and suffix along with their insert BBa_J04454 have been verified.
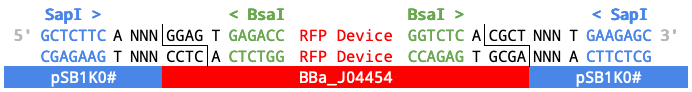
Simplified diagram of BBa_J04454 and pSB1K0# with BsaI cut site.
When four Level 0 parts are assembled into the plasmid backbone (paired with BBa_J04454) in a one-pot assembly with BsaI...

the resulting transcriptional unit (TU) will be flanked by SapI restriction sites and the specified 3 bp Fusion Sites (NNN).

Level 1 Plasmid Set
Level 1 transcriptional units in this set of plasmid backbones will be flanked by SapI recognition sites and specified 3bp fusion sites, enabling further assembly of up to 4 transcriptional units as a Level 2 Assembly.
| Registry Name | Loop Name | Fusion Site 5' | TU | Fusion Site 3' |
|---|---|---|---|---|
| pSB1K01 | pOdd1 | ATG | TU 1 | GCA |
| pSB1K02 | pOdd2 | GCA | TU 2 | TAC |
| pSB1K03 | pOdd3 | TAC | TU 3 | CAG |
| pSB1K04 | pOdd4 | CAG | TU 4 | GGT |
Level 1 -> Level 2 Assembly
Up to 4 Level 1 transcriptional units can be assembled into a Level 2 multi-transcriptional unit. By planning your assemb
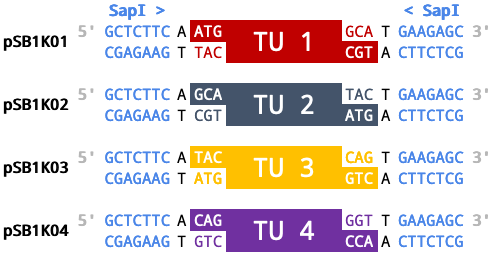
Simplified diagram of pSB1K0# set with assembled transcriptional units (TUs).
pEven Loop Vectors (for Level 2)
When four Level 1 parts (TUs) are assembled into a Multi-Transcriptional Unit (MTU) into the following plasmid backbones (with BBa_J04455), the MTU will be flanked by BsaI restriction sites and these 4 bp Fusion Sites.
| Registry Name | Loop Name | Fusion Site 5' | MTU | Fusion Site 3' |
|---|---|---|---|---|
| pSB3C01 | pEven1 | GGAG | MTU 1 | TACT |
| pSB3C02 | pEven2 | TACT | MTU 2 | AATG |
| pSB3C03 | pEven3 | AATG | MTU 3 | GCTT |
| pSB3C04 | pEven4 | GCTT | MTU 4 | CGCT |
Note:
- The documented sequence shows an additional BsaI site. This would impact assembly out of (but not into) this set of vectors. A new set of vectors should be created or used in the long term.
- Currently, Registry samples of these plasmid backbones have not been fully sequenced. Only their prefix and suffix along with their insert BBa_J04455 have been verified.
Software & Tools
Type IIS Assembly Methods
The iGEM Type IIS assembly standard is based on MoClo and Loop
- A Modular Cloning System for Standardized Assembly of Multigene Constructs
- Loop assembly: a simple and open system for recursive fabrication of DNA circuits
Accepted Standards: BioBrick RFC[10] | iGEM Type IIS RFC[1000]