Help:Assembly/Scars
- Registry Help Pages:
- TOC
- At-a-Glance
- FAQ
Accepted Standards: BioBrick RFC[10] | iGEM Type IIS RFC[1000]
Assembly Scars
A scar is a DNA sequence that is a byproduct of assembling samples of DNA parts together. As such, only composite parts are specified with scars in between subparts. Assembly systems that follow an assembly standard (BioBrick RFC10, iGEM Type IIS, etc.) create predictable scars between assembled parts.
Some assembly systems can result in "scar-less" assembly (Synthesis, Gibson, custom Type IIS), or intentionally create small spacers in between subparts.
It is important to know what scars will be in your designed composite part and how to specify scars on the Registry. This ensures that:
- the context of the subparts within your composite part are correct.
- the documented sequence of your part is accurate.
Specifying Scars
Scars are only specified on composite parts, in between subparts. After you've added a composite part, you can specify scars between subparts through the part tools menu on the "Edit sequence and features" page.
Scars are specified through square bracket "[ ]" notation, either through a shorthand or entering an exact sequence.
| Assembly System | Notation | Sequence |
|---|---|---|
| Blunt (no scar) | [0], or blank | |
| BioBrick | [10] | TACTAGAG |
| BioBrick (RBS-CDS) | [10] | TACTAG |
| User-specified | [TACT] | TACT |
Why user-specified scars?
You may have assembled the subparts together using assembly systems other than BioBrick RFC10, such as Gibson assembly, synthesis, Type IIS, etc. In some cases these may result in scars specified by the assembly system (iGEM Type IIS), in other cases there may be no scars, or scars that are unique to your assembly.
As an example, you may have completely synthesized your composite part, using parts already in the Registry. You spaced out your RBS, BBa_B0034, from your CDS, BBa_E1010, with a 6bp nucleotide sequence (Ex. TATGAC) of your choice. While this isn't truly a scar, and instead a short spacer, you should enter it as a user-specified scar:
iGEM Type IIS
For composite parts assembled with iGEM Type IIS , the scars (also known as fusion sites) are user-specified and as follows:
| Level 1 Assembly | ||||||||
|---|---|---|---|---|---|---|---|---|
| [GGAG] | Promoter | [TACT] | 5'UTR | [A]/[AATG] | *CDS | [GCTT] | Terminator | [CGCT] |
| [GGAG] | Transcriptional Unit | [CGCT] | ||||||
| Level 2 Assembly | ||||||||
|---|---|---|---|---|---|---|---|---|
| [ATG] | TU 1 | [GCA] | TU 2 | [TAC] | TU 3 | [CAG] | TU 4 | [GGT] |
| [ATG] | Multi-transcriptional Unit | [GGT] | ||||||
More information
Formation
For restriction based assembly systems like BioBrick RFC10 and Type IIS assembly, scar sequences consist of elements of the prefix and suffix after assembly. They form when the free cohesive ends of part samples ligate together.
Function
Generally, scars do not provide or code for a function, like a promoter, ribosome binding site, or terminator, so they are not considered a basic part. However, it is still important to document them correctly in the composite part editor, because scars may still have an impact, either intentional or unintentional, on your part.
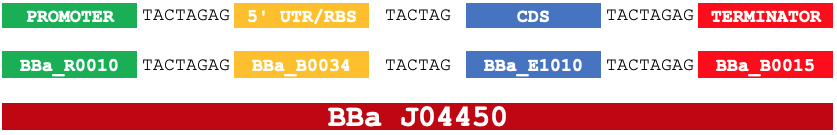
Most often, this may be as a small spacer or linker region. As an example, the 6bp scar formed when assembling BioBrick samples of the BBa_B0034 (RBS) and BBa_E1010 (CDS) results in more optimal spacing for the RBS and the start codon of the CDS, where a larger or smaller scar would cause issues with translation.
Scar sequence may also have affects on translation/transcription.
BioBrick Scars
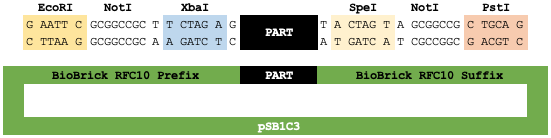
Here we have the generalized layout of a BioBrick RFC[10] part sample. The part's sequence is flanked by, but does not contain, the BioBrick prefix and suffix on the pSB1C3 plasmid backbone.
1. When assembling two BioBrick part samples (simplified), we cut the first part (BBa_R0010) with EcoRI and SpeI and the second part (BBa_B0034) with XbaI and PstI, to excise them from their respective plasmid backbones (not shown).
2. Ligating these two digested part samples together into a new plasmid backbone (not shown) allows for the cohesive ends of the cut SpeI and XbaI sites to ligate together. The resulting composite part has the BioBrick scar sequence (5' TACTAGAG 3') between the two subparts.
3. The scar sequence is a byproduct of assembly, consisting of elements of the prefix and suffix: specifically the overhang sequences ligating together, remaining bases of the original restriction enzyme recognition sites, and extra bases belonging to the prefix and suffix.
For an RBS-CDS part (why?), this scar becomes: