Part:BBa_I764001
Ethanol regulated promoter AOX1
The AOX1 promoter region from Pichia pastoris.
A complex pathway for the metabolism of methanol exists within some species of the Pichia genus. Alcohol oxidase (AO) appears to be the first and major enzyme produced in this metabolic pathway (1). Transcribed from its gene (AOX1), AO converts methanol to formaldehyde within the yeast’s peroxisome (1). A metabolic pathway for the utilization of ethanol is also present within the yeast. However, if both ethanol and methanol is present, the yeast will utilize the ethanol before consuming the methanol (2). Consequently, the AOX gene will not be expressed to produce the AO enzyme until the ethanol has been consumed.
Performance of AOX1 in in Pichia pastoris
Characterized by Beijing_4ELEVEN 2020
In our project, we choose AOX1 promoter (BBa_I764001), which is a common inducible promoter used in Pichia pastoris GS115, to control the expression. To make sure the AOX1 promoter works properly, we tested its performance with sfGFP.
We added sfGFP gene behind AOX1 promoter, inserted the whole sequence into pPIC9K backbone and transformed the plasmid into P. pastoris GS115. The recombinant strain was conducted fermentation test BMMY Medium. We measured the OD600 absorbance and fluorescence of the fermentation broth every 24 hours, and the supernatant samples during the fermentation were verified through SDS-PAGE gel electrophoresis.
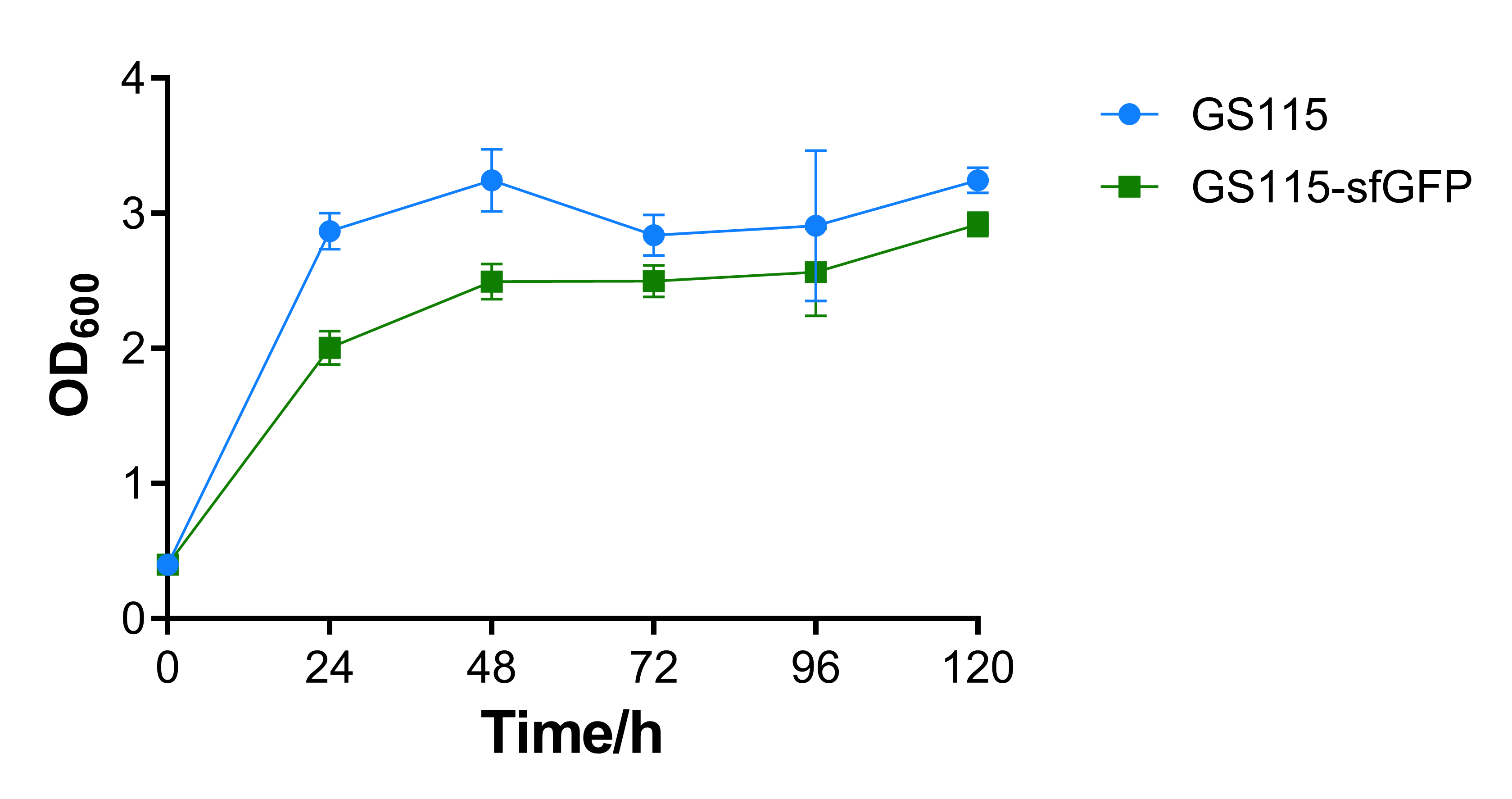
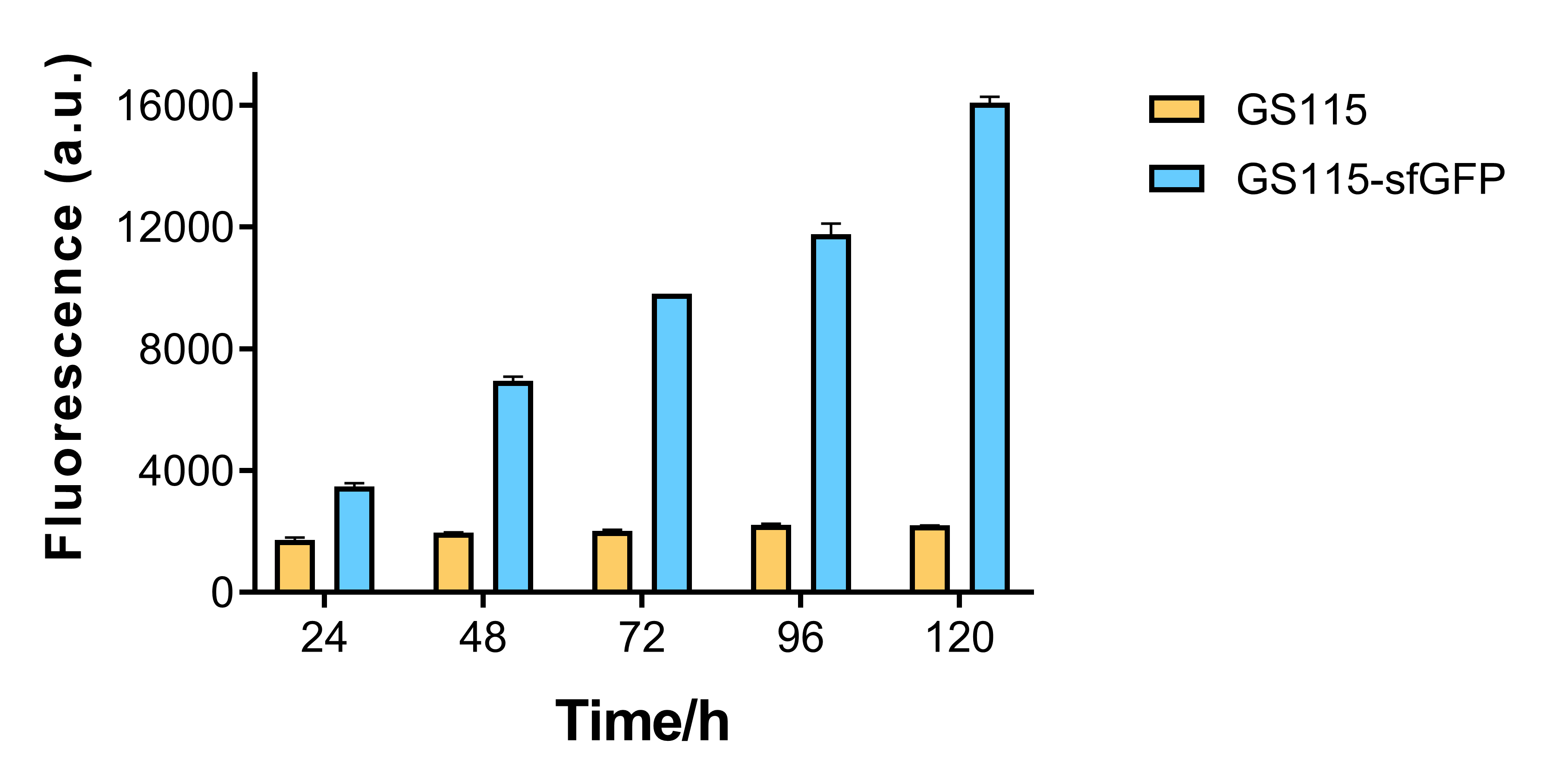
Growth curve and fluorescence test
The OD600 absorbance of the recombinant strain that contains sfGFP is a little lower than the control strain P. pastoris GS115. That may be caused by the additional expression of sfGFP, the results show that expression of heterologous gene would repress cell growth, while the repression is not intensive.
The fluorescence of recombinant strain that contains sfGFP becomes higher for every 24 hours, while that of P. pastoris GS115 remains mostly the same. This shows that AOX1 promoter under the control of methanol has the capability of promoting fluorescent protein expression.
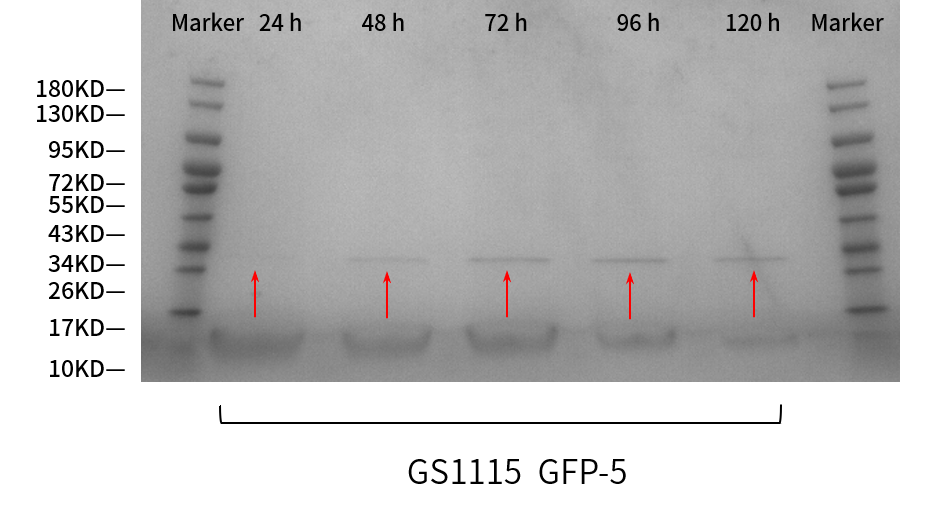
SDS-PAGE gel analysis
From the results (Figure 3), correct bands of sfGFP were shown as expected. The protein band at 24h can be barely seen; that at 48h is greater; and at 72, 96, 120h are nearly the same, all greater than at 48 h. So the clarity of the bands increases over time, which means the protein expression is getting more intensive every 24 hours.
Conclusion
All the results prove that AOX1 promoter works properly.
Source
Mycobacterium Phage Bxb1
References
Roquet, N., Soleimany, A.P., Ferris, A.C., Aaronson, S. & Lu, T.K. Synthetic recombinase-based state machines in living cells. Science 353, aad8559 (2016).
1. Cregg, James M., K. R. Madden, K. J. Barringer, G. P Thill, and C. A. Stillman. 1989. Functional Characterization of the Two Alcohol Oxidase Genes from the Yeast Pichia pastoris. Molecular and Cellular Biology. 9:1316-1323.
2. Inan Mehmet and Michael M. Meagher. The Effect of Ethanol and Acetate on Protein Expression in Pichia pastoris. 2001. Journal of Bioscience and Bioengineering. 9: 337-341.
Contribution From CPU_CHINA 2021
Group: iGEM Team CPU_CHINA 2021
Author: Peng Luo
Summary: Fluorescence intensity
Characterization from iGEM CPU_CHINA 2021
The AOX1 promoter(PAOX1) region from Pichia pastoris. It was first registered in 2007 and used as a strong promoter in Pichia pastoris. A complex pathway for the metabolism of methanol exists within some species of the Komagataella genus. Alcohol oxidase (AO) appears to be the first and major enzyme produced in this metabolic pathway. Transcribed from its gene (AOX1), AO converts methanol to formaldehyde within the yeast's peroxisome.
In order to test the function of PAOX1, we construct "PAOX1-α-factor-sfGFP-AOX1 terminator"(BBa_K3853050)(Fig. 1). If PAOX1 is functional, we can test the fluorescence intensity of sfGFP in supernatant samples obtained from the culture of recombinant P.pastoris GS115 strain.

Fig. 1 sfGFP mechanism.
1. Fluorescence intensity
Our results matched the general expected trend (Fig 2). After fermentation experiment in BMMY medium containing 0.5% methanol. The fluorescence intensity of the supernatant samples of recombinant P.pastoris GS115 containing the sfGFP gene gradually increased over time, while that of wild-type P.pastoris GS115 remained basically unchanged, which is in line with literature description[1].
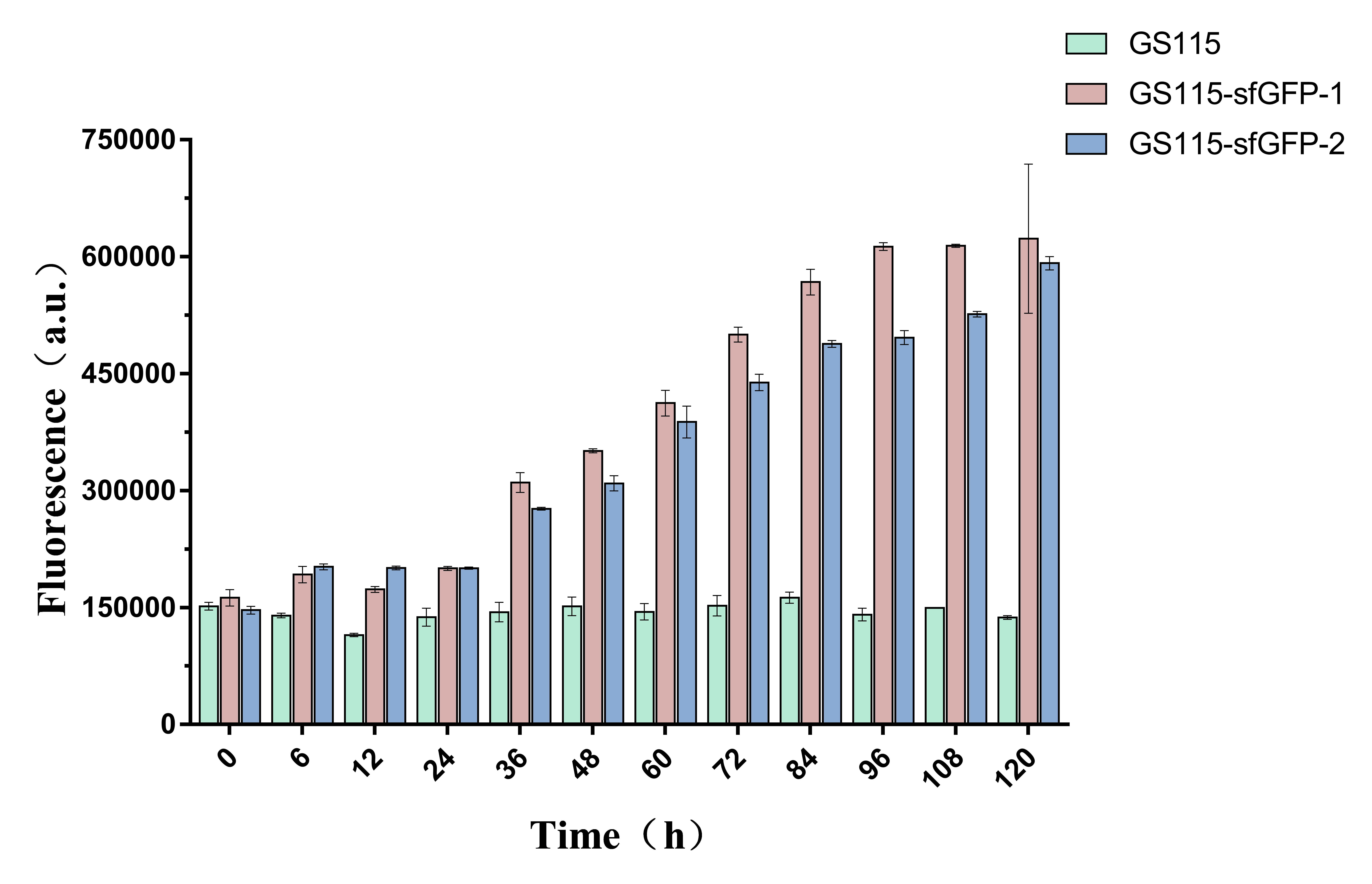
Fig. 2 Fluorescence intensity of supernatant samples obtained at different time points from the culture of wild-type P.pastoris GS115 and corresponding recombinant P.pastoris GS115 containing sfGFP gene.
2. SDS-PAGE
SDS-PAGE results (Fig. 3) also verified this phenomenon, almost no protein band before 36 h could be seen. The corresponding protein band began to appear at 36 h, and the clarity and width of the protein band gradually increased over time, which means the AOX1 promoter can continuously induce the expression of the protein.
3.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//direction/forward
//chassis/eukaryote/pichia
//promoter
//regulation/unknown
| negative_regulators | |
| positive_regulators | 1 |