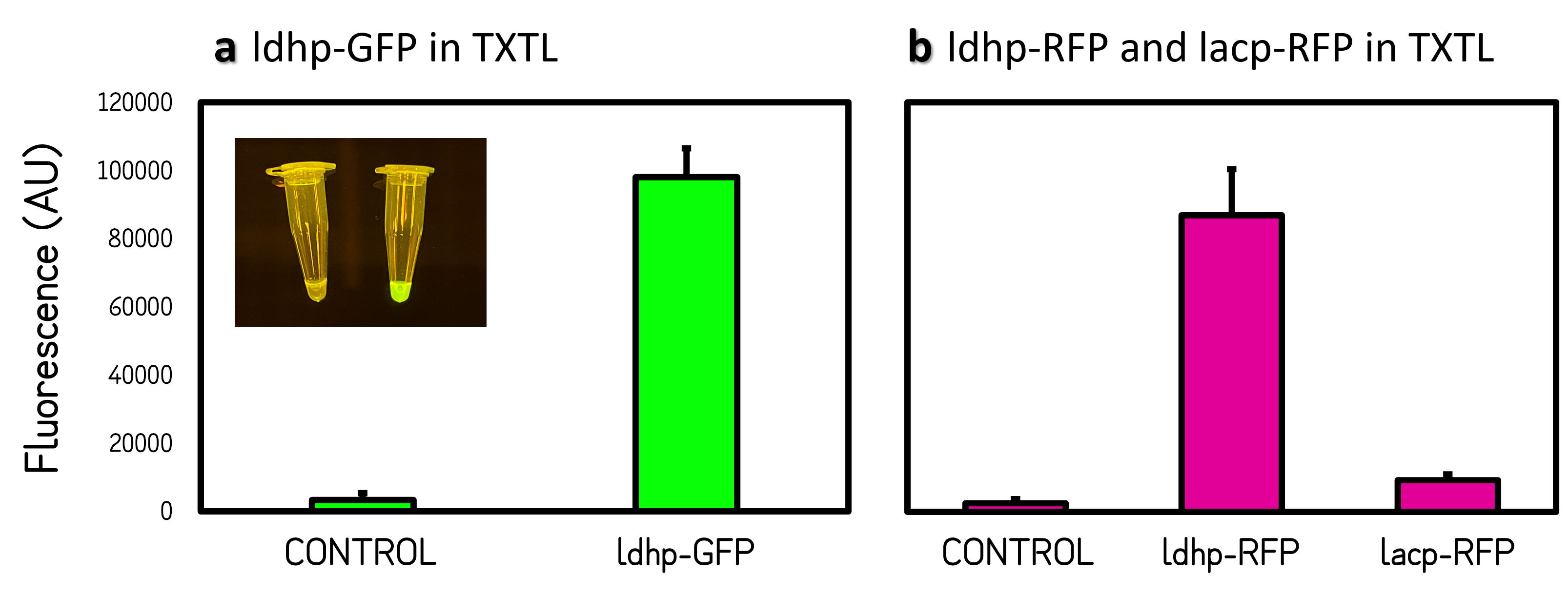Part:BBa_K3728002
pTol2: a Tol2 vector - the Tol2 transposable element
ThisTol2 transposon system is highly used in zebrafish transgenesis. The transposase protein (TPase) is from the Medaka fish (Oryzias latipes) aka Japanese rice fish, which catalyzes the transposition of the Tol2 elements through cut-and-paste mechanism. The minimal transposable Tol2 sequence (mTol2) contains 200-bp left arm and 150-bp right arm[1]. Up to 11kb DNA insert between Tol2 sequence can be integrated into the genome of nearly all vertebrates including zebrafish, frog, chicken, mouse, and human [2].
ThisA further application in synthetic biology was demonstrated by Jun Ni, et. al.[3], in which the recombinant TPase protein is fully functional in HeLa cell line and Zebrafish germline cells. In addition, the TPase can be expressed under T7 promoter in E. coli BL21 and purified with N-terminal 6xHis tag. The transposase is active in vitro and mediated the integration of DNA fragments between plasmids with Tol2 elements.
ThisIn our study, we constructed BioBrick Parts of TPase (Part:BBa_K3728000) and the BioBrick compatible Tol2 vectors (Part:BBa_K3728003) with reporter (KanR:Part:BBa_K3728004; GFP:Part:BBa_K3728005; RFP:Part:BBa_K3728006; amilCP:Part:BBa_K3728007) and Phi29 DNA polymerase genes (Part:BBa_K3728008). We prepared the In vitro transcription-translation (TXTL) system [4][5]and expressed the functional reporter proteins. The recombinant TPase and Phil29 DNA polymerase with His tag were expressed in E. coli BL21. The purified proteins were functional in the plasmid integration assay and rolling circle amplification(RCA) application, respectively.
CONSTRUCTION – BIOBRICK COMPATIBLE VECTOR
ThisMinimal Tol2 transposable element (mTol2) has been characterized that is composed of 200-bp left arm and 150-bp right arm[6]. The 19-bp to 11-kbp DNA inserts between the arms can be excised and transposed efficiently by Tol2 transposase (TPase). Therefore, we’d like to make a BioBrick compatible vector based on Tol2 mobile element (pTol2), which can be further assembled through a BioBrick standard EcoRI-XbaI-SpeI-PstI rule.
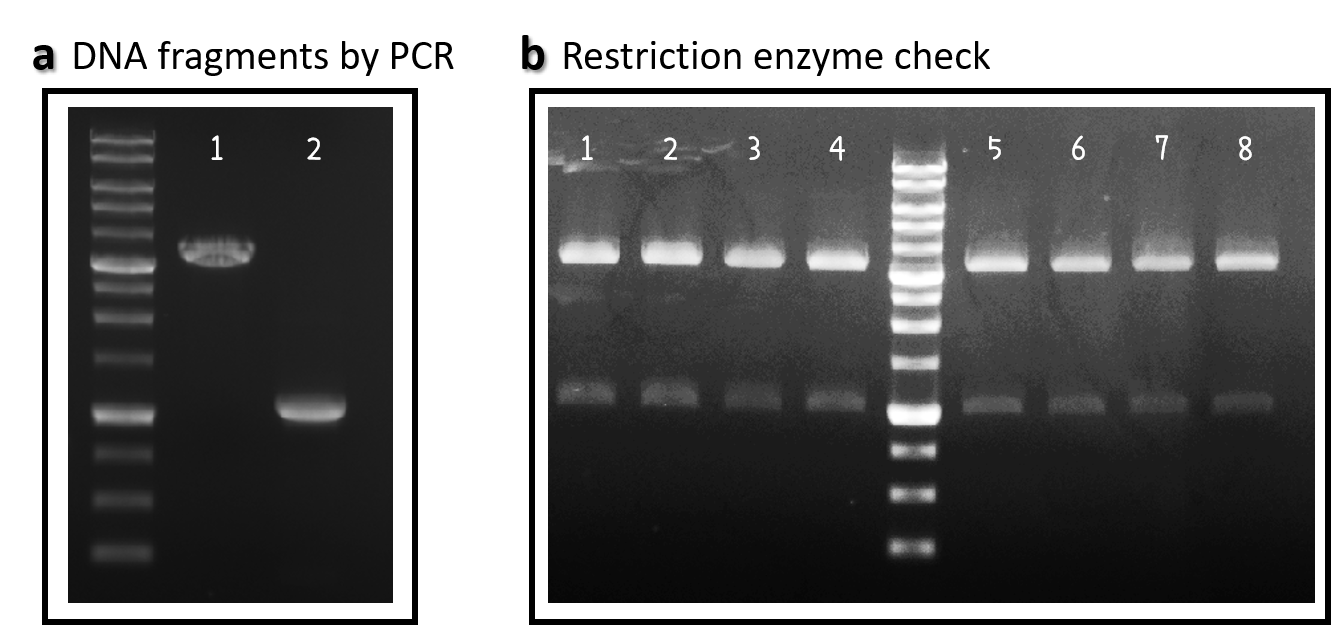
ThisWe obtained the backbone of pBSII-SK-mTol2-MCS from Addgene (Plasmid #51817), which was given by Elly Tanaka[7]. We deleted restriction enzyme sites in MCS and generated novel BioBrick Prefix (EcoRI-NotI-XbaI) and BioBrick Suffix (SpeI-NotI-PstI) elements in the both ends by PCR. The resulting DNA plasmid backbone called pTol2 (Part:BBa_K3376002) was further assembled with the Part:BBa_J04450 (i.e., the iGEM official standard insert on pSB1C3). The resulting J04450/pTol2 (Part:BBa_K3376003) was checked by PCR (Fig. 1a) and restriction enzymes (Fig. 1b) and also confirmed by sequencing.
Figure 1 | pTol2 and J04450/pTol2 constructs check. DNAs were run electrophoresis on 1% agarose gel with 1kb marker. (a) PCR producs of pTol2 (lane 1, 3429 bp) and BBa_J04450 (lane 2, 1112 bp). (b) 4 clones of J04450/pTol2 were checked by restriction enzymes (~ 3432 bp and ~1110 bp). Lanes 1-4 by EcoRI and SpeI. Lanes 5-8 by XbaI and PstI.
CHARACTERIZATION - TXTL & REPORTER ASSAY
Cell-free TXTL system
ThisIn vitro transcription and translation (TXTL) is a convenient cell-free system that has increasingly been developed to apply in synthetic biology[8][9]. In addition to achieve biosafety level, TXTL becomes powerful in prototype characterization of genetic parts, devices and circuits. Moreover, TXTL is particularly useful to express and purify proteins which are toxic, insoluble or unstable in cell-based system. Furthermore and amazingly, Dr. Vincent Noireau’s lab has demonstrated cell-free TXTL application in infectious bacteriophage production, in which T7 phage (40kbp, 77 genes, dsDNA) and T4 phage (170kbp, 289 genes, dsDNA) genome replication, synthesis, assembly can be performed in vitro just in a single test tube[10][11]
Promoter Activity
ThisTo test the TPase activity and Tol2 transposon system, we inserted a kanamycin resistance gene (KanR) cassette (Part:BBa_K3376004) and the reporters of ldhp-GFP-Tr(Part:BBa_K3376005), ldhp-RFP-Tr(Part:BBa_K3376006) and ldhp-amilCP-Tr(Part:BBa_K3376007) between the transposable elements on the pTol2 vector. ldhp is a constitutive and broad-host-range promoter, which was originally cloned and driving the lactate dehydrogenase gene in S. mutans. We have characterized the ldhp activities in S. mutans and E. coli in our project of iGEM 2020, as well as Salmonella and TXTL in this project of iGEM 2021.(Part:BBa_K3376000)
ThisThe GFP and RFP fluorescence intensities driven by ldhp on pTol2 vectors were measured at high level in TXTL reaction (Fig. 2). The strong GFP fluorescence can even be visualized by naked eyes under a Blue LED Illuminator. Compared the activities of ldhp to lac promoter (lacp), lacp is inhibited in TXTL because the extracts of E. coli Rosetta 2 (DE3) contains LacI repressor, which can be relieved by IPTG induction or using E. coli DH5α as extracts.
Figure 2 | Promoter activities on pTol2 vector in TXTL. GFP fluorescence was measured at Ex/Em = 500/530 nm using a microplate reader of BioTek Synergy H1. RFP was at Ex/Em = 586/611 nm. KanR/pTol2 in TXTX was set as a background control. AU means arbitrary unit. (a) ldhp-GFP-Tr/pTol2 activity in TXTL. The inset photo was captured under a blue LED light. (b) ldhp-RFP-Tr/pTol2 and J04450/pTol2 (i.e., lacp-RFP-Tr) in TXTL.
CHARACTERIZATION – PLASMID INTEGRATION
In vitro Integration Assay
ThisIn vitro integration assay was used by Jun Ni, et al. to characterize the activity of purified recombinant Tol2 transposase (TPase) and the transposition of Tol2 mobile element[10]. We prepared the purified TPase from TXTL (Fig. 2) and performed PCR to generate KanR, ldhp-GFP-Tr and ldhp-amilCP-Tr (expressing blue color) DNA fragments flanked by 200-bp right and 150-bp left arms of pTol2 (Part:BBa_K3728002). The mixtures of TPase, Tol2 mobile inserts and a target plasmid of pSB1C3 were incubated at 30°C for 2 hours. The resulting DNAs were cleaned up and subjected to transform E. coli DH5α competent cells. The colonies displaying kanamycin resistance, green fluorescence or blue color were counted as successful jumping to plasmids by active purified TPase. And the integration rate was calculated by comparing with chloramphenicol resistance or red colonies from pSB1C3 backbone carrying Part:BBa_J04450 part (i.e., RFP coding device).
ThisGFP/Tol2-integrated plasmid can transform E. coli to exhibit weak to strong green fluorescence in Fig. 3. Two plasmids of GFP-positive bacteria were extracted and checked by restriction enzymes. They are larger than pSB1C3 when single cut on the backbone by ApaLI (Fig. 4b). The schematic map of Fig. 4a showed the possible position of integration by a BamHI-cut on the insert and a ApaLI-cut on the backbone (Fig. 4c). The rate of successful integration was calculated by the ratio of numbers of KanR, GFP and BLUE colonies to CmR or RED colonies, respectively (Fig. 4d). The ratio was between 0.2% to 0.9%, of which data are consistent with the observation by Jun Ni, et al[10]. In sum, we can modify plasmid DNAs in vitro with an insert between Tol2 mobile elements (DONOR) and purified TPase enzymes (HELPER) from TXTL reaction.
- Figure 3 | E. coli colonies on Cm agar plates were transformed by the mixture of GFP/Tol2 and pSB1C3 with TPase or without TPase as a control.
- Figure 4 | Possible integration map and ratio. (a) Schematic maps showed the predicted integration sites. (b, c) pSB1C3::GFP/Tol2 Clone #1 (lane 1) and #2 (lane 2) or pSB1C3 as a control (lane 3) were cut by ApaLI on the backbone (b) or cut by ApaLI with a BamHI cut on the insert (c). DNA was analyzed by electrophoresis on 1% agarose gel with a 1kb marker. (d) The successful integration ratios are calculated by the numbers of colonies of pSB1C3::KanR/Tol2 on Kan agar plate divided by those of pSBC13 (CmR) on Cm agar plates or by the numbers of pSB1C3::GFP or pSB1C3::BLUE divided by colony numbers of pSB1C3 (RED) on Cm agar plates such as shown in Figure 3.
APPLICATION – PHAGE ENGINEERING
Construction - ldhp-Phi29 DNA pol-Tr/pTol2
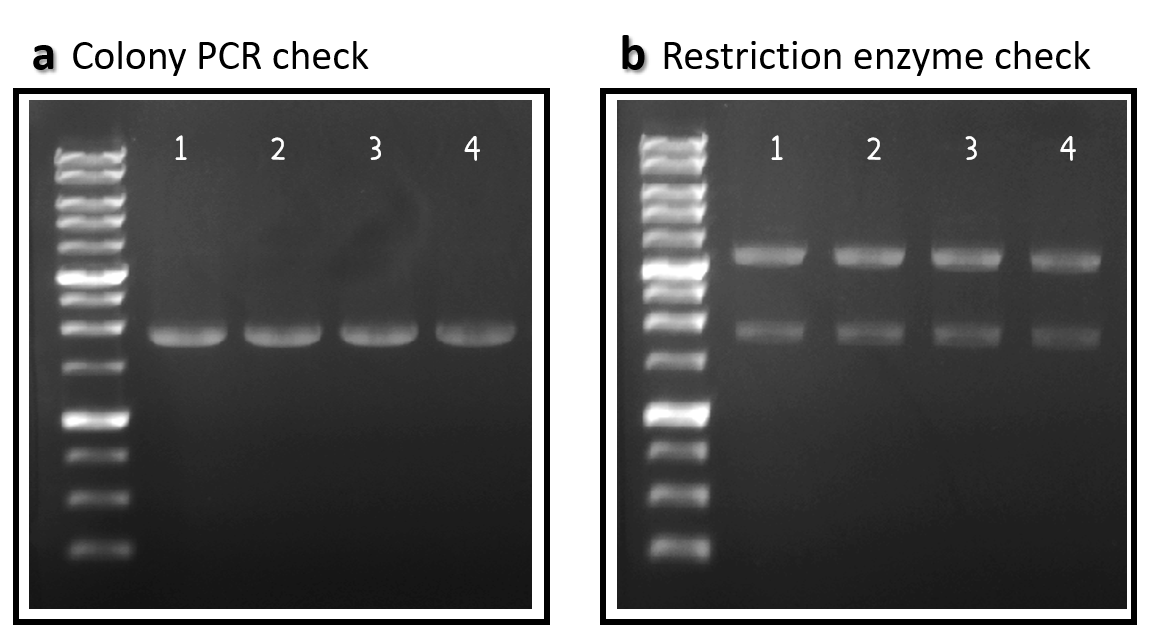
ThisTo engineer RCA reporter Salmonella phage, we requested Part:BBa_K3352001 (Φ29 DNA Polymerase with His-Tag and GS linker Sequence) from iGEM team TAS_Taipei in 2020. The part was assembled with ldhp promoter Part:BBa_K3376000 and a double terminator Part:BBa_B0015 onto the Tol2 mobile element vector (pTol2, Part:BBa_K3728002). The resulting composite part named ldhp-Phi29 DNA pol-Tr/pTol2 (Part:BBa_K3728008) was checked by colony PCR, restriction enzymes and sequencing (Fig. 2).
Figure 2 | ldhp-Phi29 DNA pol-Tr/pTol2 construct check. DNAs were run electrophoresis on 1% agarose gel with 1kb marker. (a) 4 colonies were subject to PCR with ldhp forward primer and a reverse primer in the end of Phi29 gene (PCR product size: 1965 bp). (b) The DNAs were extracted and digested by EcoRI and PstI (3573 and 1973 bps).
Protein Expression & Functional Assay - Ф29 DNA Polymerase
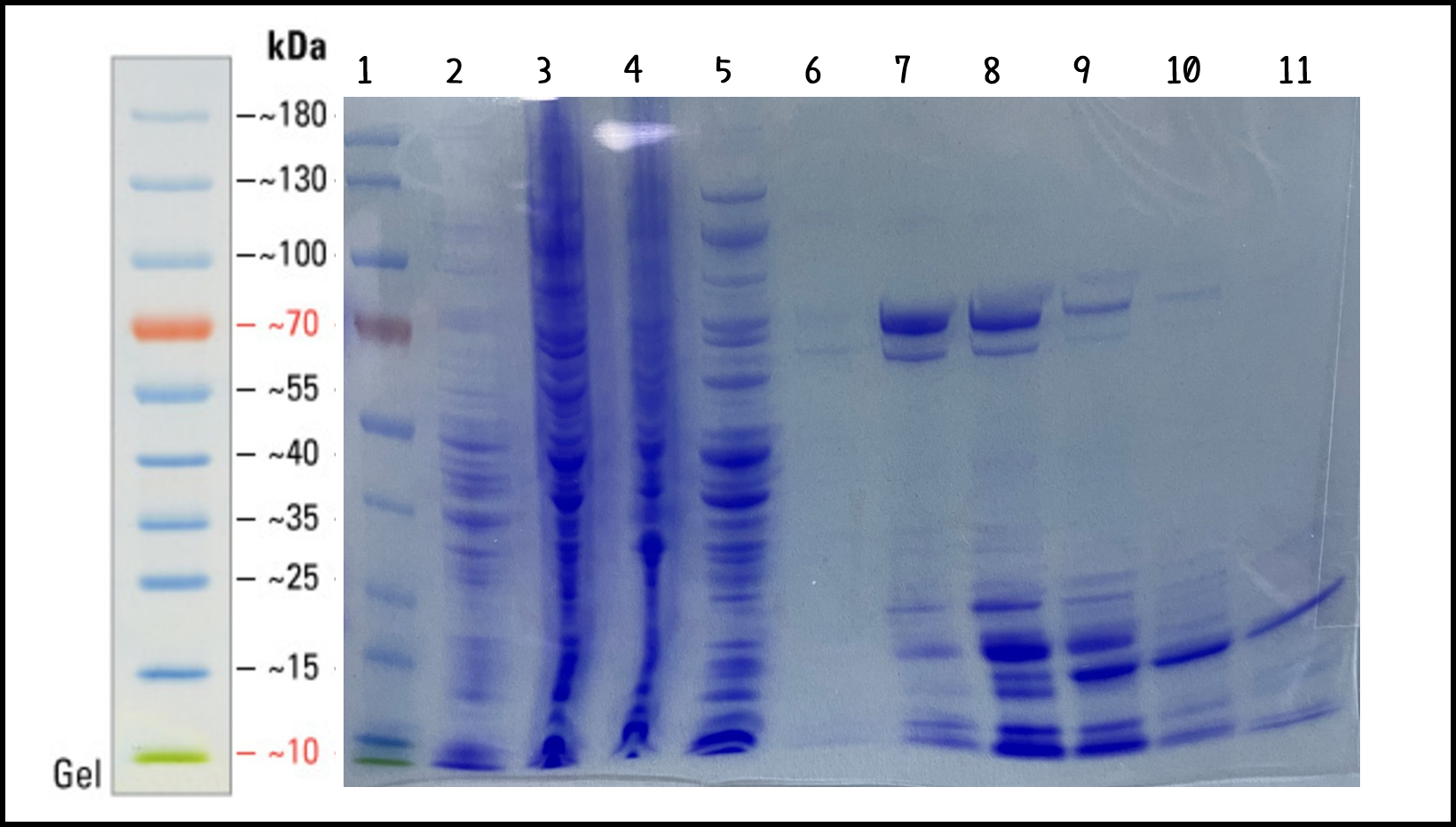
ThisTo characterize the function of recombinant phi29 DNA polymerase, TXTL cell-free system can solve the problem caused by the difficulty in bacterial transformation or without suitable plasmid vectors. We took the DNA of ldhp-Phi29 DNA pol-Tr/pTol2(Part:BBa K3728008)mixed into TXTL reaction with Salmonella extracts. The recombinant His-tagged phi29 DNA polymerase protein was purified through Nickel column and analyzed by SDS-PAGE and Coomassie Blue Staining (Fig. 4). The isolated proteins in Elution #4 and #5 were at the size of around 70 kDa as predicted (His-phi29 DNA polymerase: 590 amino acids, 68 kDa) and collected for the following studies.
Figure 4 | His-phi29 DNA polymerase was expressed in TXTL using Salmonella extracts and purified by Nickel column. 5 μg of protein lysates were analyzed by SDS-PAGE and Coomassie Blue Staining using 4–12% gradient gel (NuPAGE™, Thermo Fisher Scientific Inc.) Lane: (1) PageRuler™ Prestained Protein Ladder, (2) Salmonella cell extracts (no DNA control), (3) total lysates in TXTL, (4) flow-through, (5) wash-through, (6) Elution #3, (7) Elution #4, (8) Elution #5, (9) Elution #6, (10) Elution #7, (11) Elution #8.
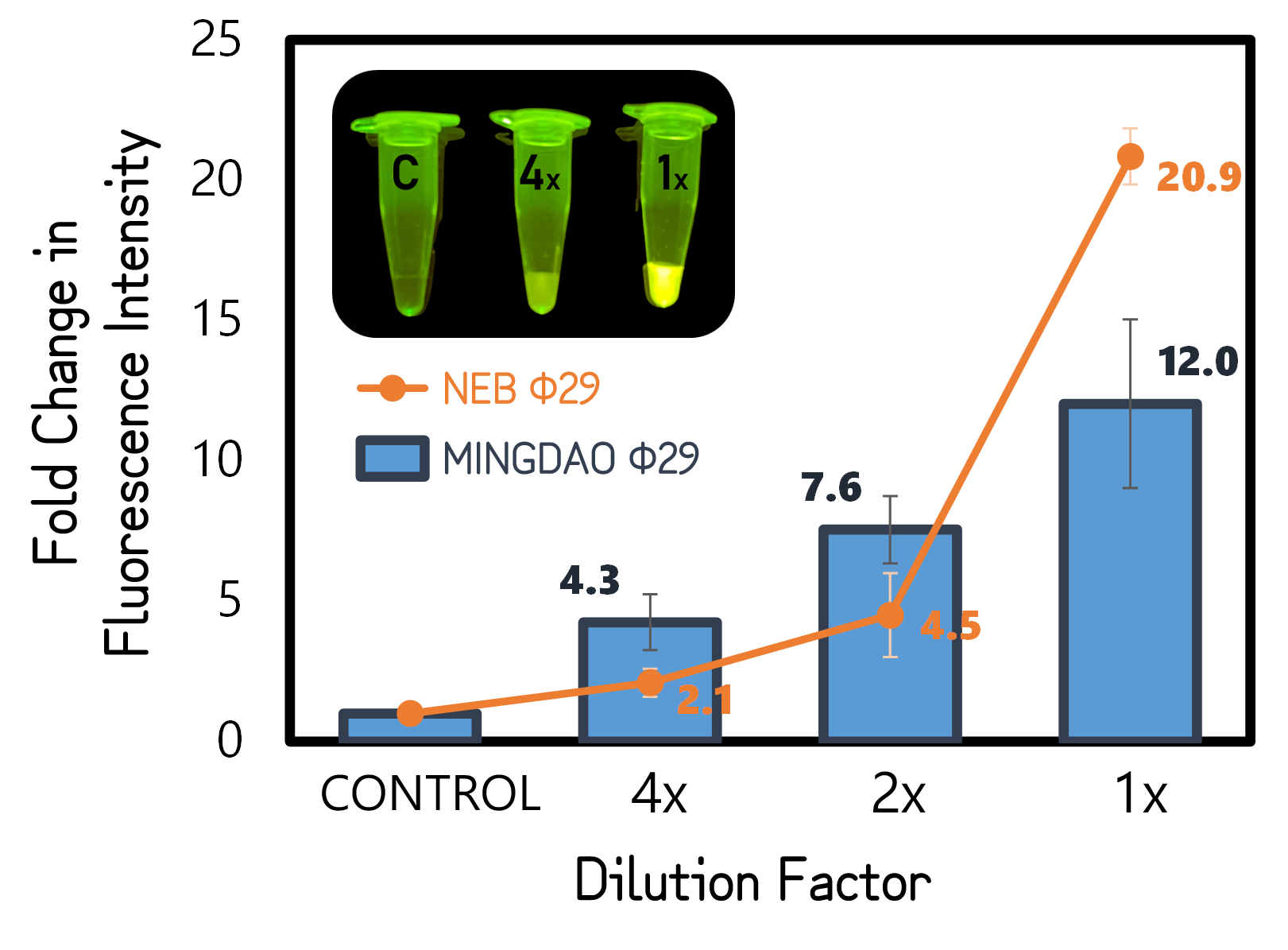
ThisTo test functionality of phi29 DNA polymerase, we performed RCA by mixing a circular ssDNA and primer in the buffer with our purified phi29 DNA polymerase (Φ29) from TXTL using Salmonella extracts or a commercial recombinant Φ29 from New England Biolabs Inc. (NEB) as a positive control. The Φ29 enzymes were diluted with various factors in the assay. After incubation at 30°C for 1 hour, the RCA products were stained with EvaGreen DNA dye and subjected to a microplate reader to measure signals at Ex/Em=500/530 nm. And the fold changes in fluorescence intensity were calculated by dividing the values from Φ29-untreated groups (as controls). As shown in Fig. 11, a 12-fold change was achieved with our Φ29, indicating the functionality and the activity of ldhp-Phi29 DNA pol-Tr/pTol2 that are comparable to the commercial NEB Φ29 enzymes. Moreover, the green fluorescence can readily be seen with a blue led light, even using a 4 times diluted Φ29, proving the super high processivity of phi29 DNA polymerase in DNA amplification.
Figure 5 | RCA assay using TXTL-expressed (MINGDAO) or commercial (NEB) phi29 DNA polymerase (Φ29). The commercial Φ29 was purchased from NEB with a defined activity by units. The control was set without Φ29 treatment. The concentration and dilutions of enzymes were, respectively, 4, 2, 1 g/l for MINGDAO Φ29 and 1, 0.5, 0.25 units for NEB Φ29. The EvaGreen DNA binding signals were read at Ex/Em=500/530 nm in BioTek Synergy H1 Microplate Reader.
Reference
- ↑ Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006 Oct;174(2):639-49. doi: 10.1534/genetics.106.060244.
- ↑ Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8 Suppl 1(Suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7
- ↑ Ni J, Wangensteen KJ, Nelsen D, Balciunas D, Skuster KJ, Urban MD, Ekker SC. Active recombinant Tol2 transposase for gene transfer and gene discovery applications. Mob DNA. 2016 Mar 31;7:6. doi: 10.1186/s13100-016-0062-z.
- ↑ Garenne D, Noireaux V. Cell-free transcription-translation: engineering biology from the nanometer to the millimeter scale. Curr Opin Biotechnol. 2019 Aug;58:19-27. doi: 10.1016/j.copbio.2018.10.007.
- ↑ Rustad M, Eastlund A, Marshall R, Jardine P, Noireaux V. Synthesis of Infectious Bacteriophages in an E. coli-based Cell-free Expression System. J Vis Exp. 2017 Aug 17;(126):56144. doi: 10.3791/56144.
- ↑ Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006 Oct;174(2):639-49. doi: 10.1534/genetics.106.060244.
- ↑ Khattak S, Murawala P, Andreas H, Kappert V, Schuez M, Sandoval-Guzmán T, Crawford K, Tanaka EM. Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nat Protoc. 2014 Mar;9(3):529-40. doi: 0.1038/nprot.2014.040.
- ↑ Marshall R, Noireaux V. Synthetic Biology with an All E. coli TXTL System: Quantitative Characterization of Regulatory Elements and Gene Circuits. Methods Mol Biol. 2018;1772:61-93. doi: 10.1007/978-1-4939-7795-6_4.
- ↑ Tinafar A, Jaenes K, Pardee K. Synthetic Biology Goes Cell-Free. BMC Biol. 2019 Aug 8;17(1):64. doi: 10.1186/s12915-019-0685-x.
- ↑ 10.0 10.1 10.2 Shin J, Jardine P, Noireaux V. Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction. ACS Synth Biol. 2012 Sep 21;1(9):408-13. doi: 10.1021/sb300049p.
- ↑ Rustad M, Eastlund A, Jardine P, Noireaux V. Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction. Synth Biol (Oxf). 2018 Jan 22;3(1):ysy002. doi: 10.1093/synbio/ysy002.
Note: The map was generated and sponsored by SnapGene.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3452
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 29
Illegal NotI site found at 3458 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3452
Illegal BglII site found at 3340
Illegal XhoI site found at 3419 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 3452
Illegal suffix found at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 3452
Plasmid lacks a suffix.
Illegal XbaI site found at 3467
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NgoMIV site found at 642 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal BsaI site found at 1818
Illegal SapI.rc site found at 2900
| None |