Part:BBa_K1075032
ccdA - Antitoxin in ccdA/ccdB system
CcdA inhibits the toxin ccdB by binding to it and thus represses cell death . There is no stop codon because we used it in fusion with the E.Coli ssrA(DAS+4) tag.
Usage and Biology
The ccd module is a toxin-antitoxin (TA) system. The module naturally occurs on the F Plasmid in Escherichia coli bacteria and is essential for their survival. Normally the toxin ccdB is inactivated by the presence of the antitoxin ccdA in the form of a ccdAB complex. If ccdA is no longer available, ccdB inhibits DNA gyrase which leads to cell death. Gyrase is a type IIA topoisomerase specific to E. coli and is able to produce negative DNA supercoiling by making a double-strand break in the DNA and religating it. CcdB stabilizes the gyrase cleavage complex and thus blocks the catalytic function of the gyrase. That means that the gyrase remains bound to the DNA and the cleaved DNA is not religated. DNA- and RNA polymerases can’t copy the DNA anymore and cell proliferation as well as protein biosynthesis is stopped. The double-stranded breaks in the DNA initiate cell death. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1635281/][http://www.ncbi.nlm.nih.gov/pubmed/?term=A+Common+Origin+for+the+Bacterial+Toxin-Antitoxin+Systems+parD+and+ccd]
Characterization
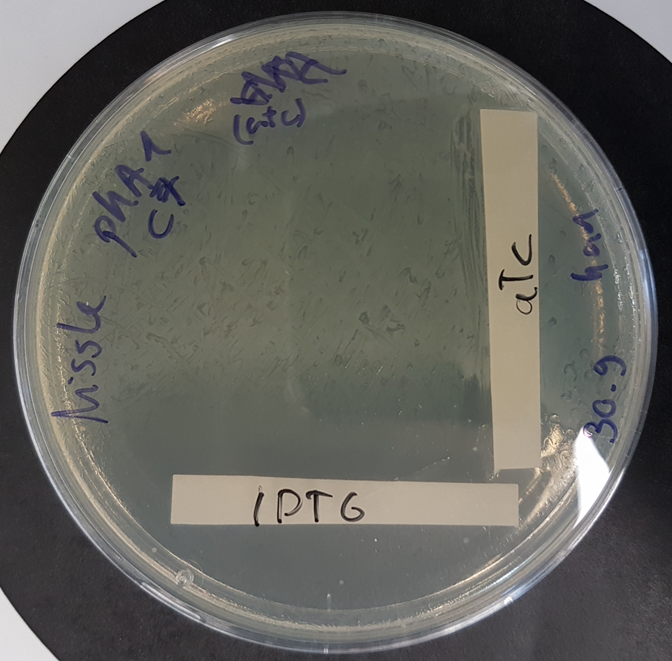
Contribution of the Igem Team UNILausanne 2020 Functional kill switch assay with IPTG and aTc gradients on agar plate
E.coli Nissle 1917 were transformed with a plasmid containing the part BBa K3482016 and the part BBa_K3482020 (CcdA antitoxin part) and plated with a gradient of aTc and IPTG on agar plate. The plate shows strong activity of the CcdA antitoxin with aTc induction, whereas IPTG induction promotes production of the CcdB toxin, resulting in a number of surviving cells (probable mutants) proportional to the dilution of the plated culture.
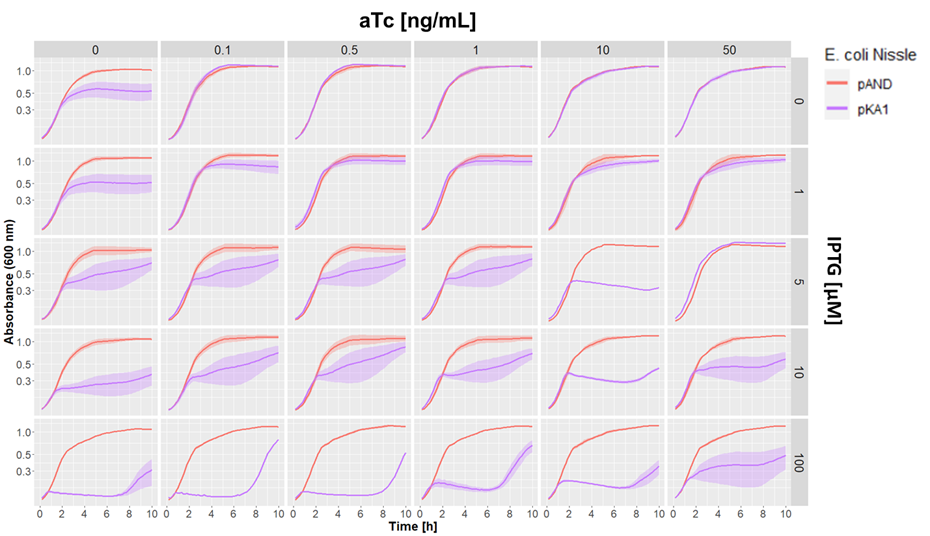
We aslo tested our pKA1 plasmid encoding for the ccdB toxin and ccdA antitoxin in E. coli Nissle 1917 ΔclbA. We used different concentrations of IPTG and aTc to study the effect of differential expression of the toxin and antitoxin on the growth of our strain. We compared this to the strain transformed with the empty vector pAND
Dose-response growth curve of E. coli Nissle 1917 ΔclbA harboring kill switch plasmid pKA1 at 37°C. E. coli Nissle with pAND (red line), E. coli Nissle with pKA1 (blue line). The lines and shade represent the mean ± standard error.
We observed desired growth inhibition of the strain with pKA1 at high IPTG and low aTc concentrations, while the pAND strain showed no alteration in growth in any of the tested conditions. The uniformity of these growth curves of the strain with pAND demonstrates that the standardized culturing and OD normalization of our protocol can yield highly reproducible results.
We saw an effect of IPTG on growth inhibition starting from 5 µM IPTG. At maximum IPTG concentration, we observed complete absence of growth of the pKA1 strain for the first 7 h. However, afterwards, we observed that the bacteria were able to grow. This was probably due to mutations in the kill switch system. In contrast, at lower IPTG concentrations (5 and 10 µM), we still observed an inhibitor effect on growth, which albeit being less strong, was more constant and persisted for at least 10 h. This suggests that decreasing the expression of the toxin lowers the selective pressure on the kill switch system, which makes this configuration more evolutionarily stable. In fact, while we observe a strong uninhibited growth of escape mutants after 7 h with 100 µM IPTG, at 10 µM IPTG the cells show a constant repression on their growth rate, which indicates that no or little escape mutants have emerged.
Overall, we demonstrated that the ccdB/ccdA encoding pKA1 plasmid could efficiently inhibit the growth of E. coli Nissle 1917 at 100 µM IPTG for up to 7 h, and provided even longer growth inhibition at 5 and 10 µM IPTG. sequence and features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |