Part:BBa_J23119:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_J23119
Experience of the IIT_Madras iGEM 2010 team.
As submitted on 29th Oct 2010, 0350 IST (+0530 GMT)The iGEM 2010 IIT Madras team worked with J23119 as a reference in order to characterise a new pH inducible promoter that we have added to the registry this year. While analysing the results of the charactersation experiment we conducted we found that this promoter, in conjunction with an RBS (J23119 + B0034) has upregulated activity at low pH (4.5 - 5).
Experimental Design
The experiment we conducted involved the use of the construct J23119+B0034 in the plasmid pGL3Basic, which is a promoter-less plasmid that contains Firefly Luciferase as a reporter. The plasmid was transformed into DH5Alpha and cultured as a primary inoculum in Luria Bertani media. Once the inoculum reached an OD600 of 1.0 the culture was used to inoculate pH adjusted media of pH 4.5, 5, 5.5, 6 and 7, all in duplicates. We regularly measured OD600 for these 10 cultures until their OD600 reached 0.5, or reached saturation. We then took 3 independent samples of these cultures with vigorous shaking in-between consecutive samplings.Observations and Inferences
In this way we obtained a measure of the Luminescence of 30 samples in an arbitrary unit, RLU/s (Relative Luminescence units per sec). The data is as tabulated below.
Table 1 and 2: Data representing experiment done at various pH in duplicate and sampling done in triplicate. The Luminescence has been normalised using harvest-time OD600 readings of each culture.
The data was also plotted, as shown below.

Figure 1: Experimental data plotted for the duplicates showing high level of reproducibility in the data.
From these results we infer that it is highly possible for the constitutive promoter J23119 to be under the control of another stress factor based on pH. In this case it is possible that due to the high pH stress in the medium, the luciferase is over-expressed. It should also be noted that our pH inducible (in the range of 5 - 6.5) did not show this increased activity at low pH.
For more information on our contribution to the world of Synthetic Biology, check out our wiki.
References
- pGL3Basic vector backbone, as distributed by Promega. http://www.ncbi.nlm.nih.gov/nuccore/13195703, last accessed on 30th Oct 2010, 0350 IST (GMT +0530).
- DH5Alpha E.coli strain. http://ecoliwiki.net/colipedia/index.php/DH5_alpha, last accessed on 30th Oct 2010, 0350 IST (GMT +530).
Experience of CLSB-UK-2016 iGEM team with Part:BBa_K2078002
Experimental Design
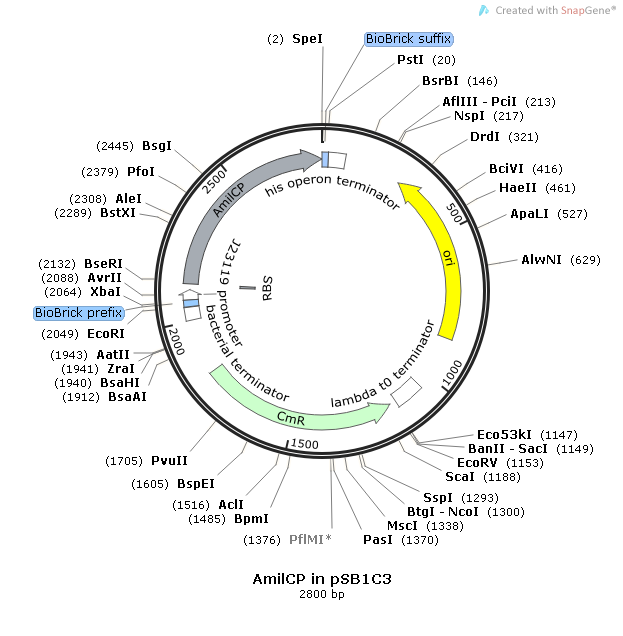
This consensus promoter was assembled with the reporter protein, blue-purple chromoprotein AmilCP. The construct was carried on the pSB1C3 plasmid for expression and amplification in E-Coli. The plasmid also contained chloramphenicol acetyltransferase for resistance during plating (CmR).
Observations and Inferences
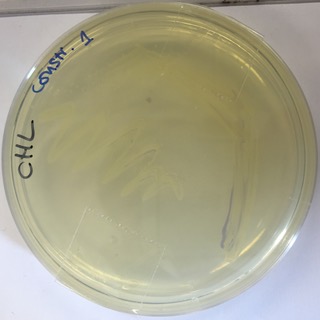
Our E.coli grew slower than other transformants and we were also unable to recover as much plasmid by miniprep. The use of this strong consensus promoter slows down expression times of the AmilCP Part:BBa_K592025 in E-Coli by around 24hours. This image shows a pellet from 5ml LB broth with transformed E.coli and confirms that the part is expressed, but the amount of pellet was much less than we would normally expect to get from 5ml.
The delay in E-Coli growth is also shown by this 24hour streak plate. The chromoprotein is only just becoming visible suggesting that such a strong consensus sequence slows down both the expression time of the part and growth time of E-Coli.
.
User Reviews
UNIQdd864bf300584a80-partinfo-00000001-QINU
|
No review score entered. Sriram Srikant |
We used j23119 along with the RBS B0034 to provide a constitutive baseline expression during our characterization of p170 promoter cassette. To our suprise we found that under variable ph conditions the expression of the promoter varied, it showed extreme upregulation at low ph (~4.5). |
|
•••••
[http://2011.igem.org/Team:KULeuven K.U.Leuven iGEM 2011 Team] |
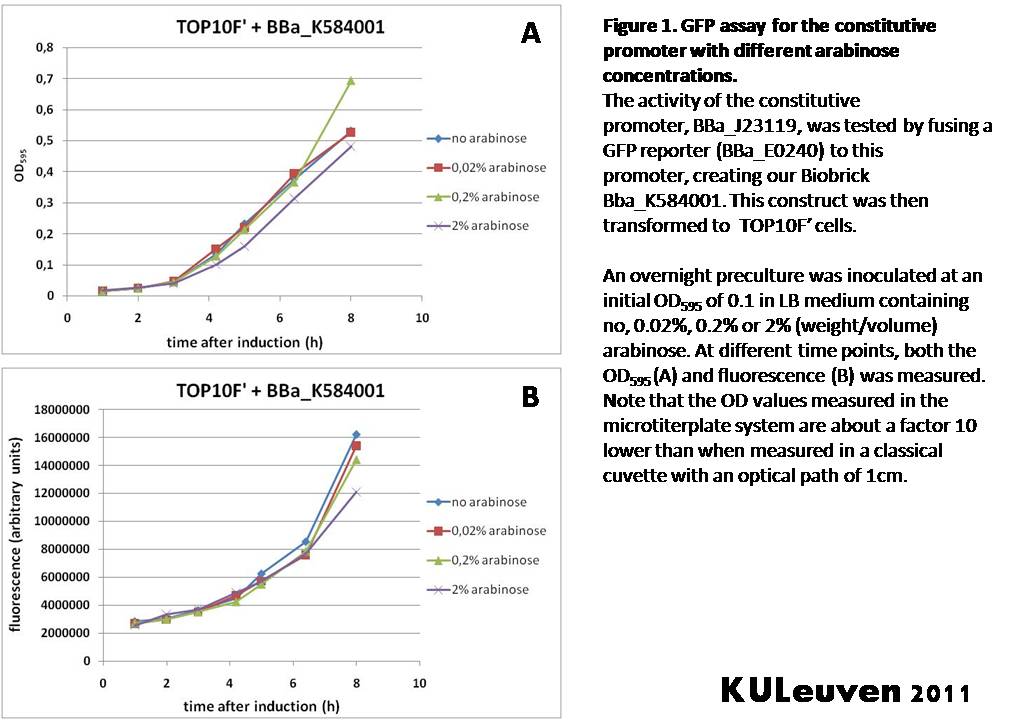
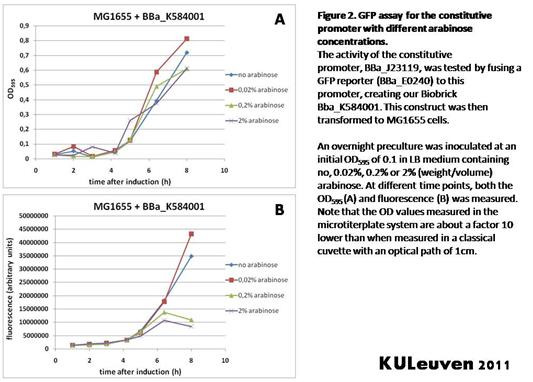
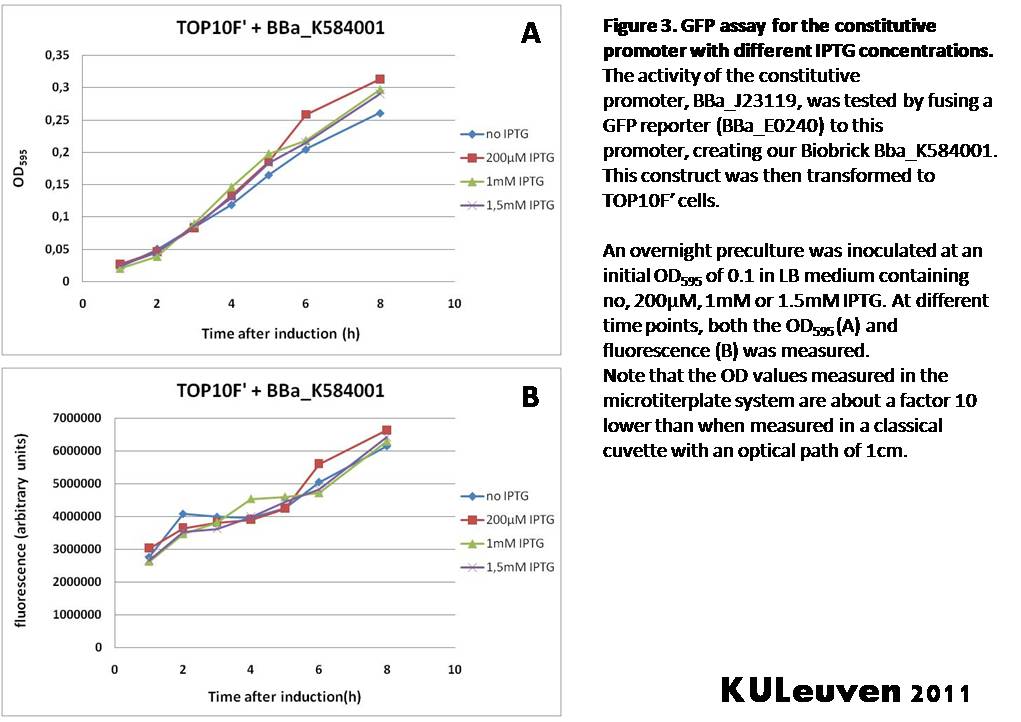
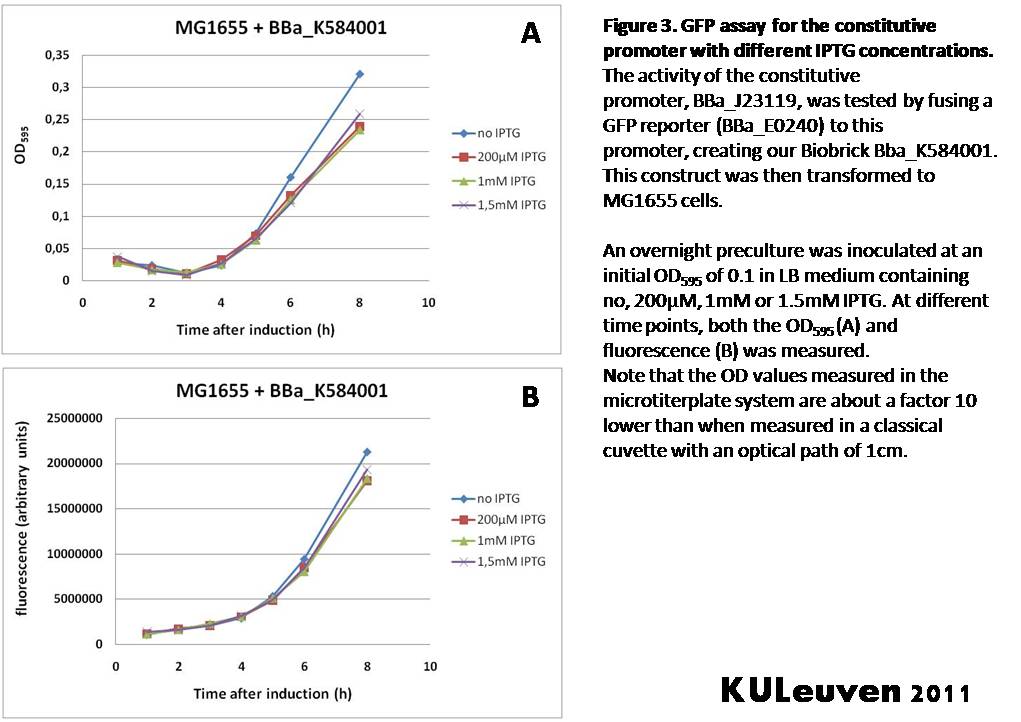
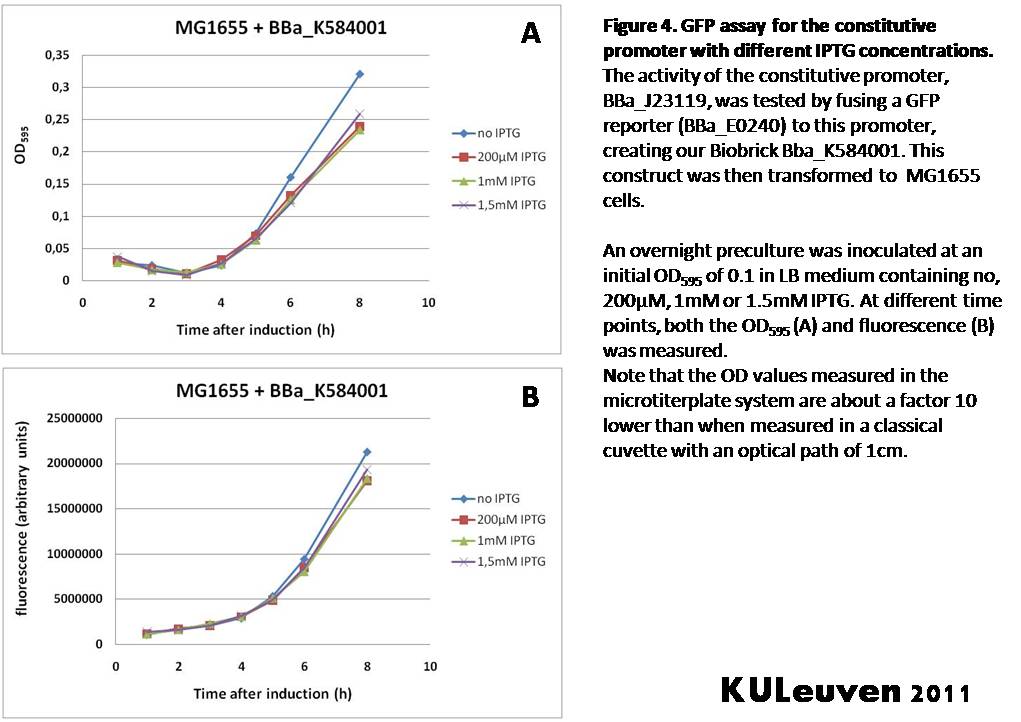
Characterization by K.U.Leuven 2011 iGEM Team We fused the constitutive promoter to a GFP reporter, and assayed the promoter’s activity after addition of different amounts of arabinose and IPTG. As such these results can serve as a control for the results obtained for the arabinose-inducible BBa_K584000 and IPTG-inducible BBa_K584002 bricks. We tested the activity in a TOP10F’ (Figure 1 & 3) as well as a MG1655 (Figure 2 & 4) E.coli strain background. For more information on E.coli strain descriptions, we recommend the following website: [http://openwetware.org/wiki/E._coli_genotypes E.Coli_Genotypes]. Apart from a small growth defect and a somewhat lower fluorescence at the 8 hour time point after the addition of 2% arabinose, the activity of the constitutive promoter in TOP10F’ cells is unaffected by arabinose addition, as seen in Figure 1. For the MG1655 cells (Figure 2), however, we see clear effects of adding arabinose at concentrations of 0.2% or 2%, with promoter activities significantly lower than the control condition. This points to interference of high inducer levels with the activity of the promoter and the importance of metabolism of the inducer, as arabinose can be metabolized by MG1655 cells, but not by TOP10F’ cells. For both TOP10F’ and MG1655 cells, the activity of the constitutive promoter is unaffected by the addition of IPTG (Figures 3 and 4).
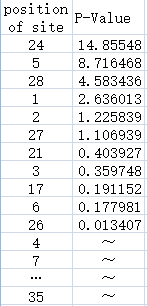
Experience of NAU-CHINA-2016 iGEM teamExperimental DesignTo evaluate each site of the Anderson Promoter`s effect on the whole promoter, the NAU-CHINA Team used TASSEL5 to analyse the correlation degree between each mutation and the strength of the promoter. Error creating thumbnail: File missing The figure shows the correlation degree between the each position of the site of the promoter and the strength of the promoter using a series of Anderson promoters to analyse. The abscissa of the figure represents the position of the site of the sequence of the promoter. The position start at "0", so the last of the number of the site is "34". The ordinate of the figure represents the P-Value of the each site. The higher the P-Value is the higher effect of the site is,and the lower strength of the promoter is at the same time. For example ,we will find out that the 24th site reduce the strength of the promoter greatly from the figure. |

 1 Registry Star
1 Registry Star