Part:BBa_K1065102
Wintergreen device: Plac + PchBA + AraCpBAD + BSMT1
This was the device employed in the project B. Fruity to inhibit the ripening maturation with the production of Methyl Salicylate by the UNITN-Trento 2013 iGEM team. The device includes the enzyme PCHBA under the control of pLac promoter and BSMT1 under the control of an arabinose inducible promoter [2].
Usage and Biology
The chorismate, a metabolic intermediate of the Shikimate pathway which many plants and bacteria have, undergoes first a reaction of isomerization by the isochorismate synthase PchA. Salicylate is then obtained by the action of PchB an isochorismate pyruvate lyase. Both enzymes are from the micro-organism Pseudomonas aeruginosa [1] . In the final part of the reaction, BSMT1, a methyltransferase, transfers a methyl group from the S-adenosyl-L-methionine synthesized by the SAM synthetase (in E. coli there's already a basal expression of the enzyme).
To improve methyl salicylate production, iGEM UniTN Trento 2013 created another device containing also SAM synthetase: BBa_K1065106.

Characterization
MeSA detection
MeSA is an highly volatile liquid with a distinct minty fragrance. We exploited the physical properties of MeSA to quantify its production by gas chromatography using a Finnigan Trace GC ULTRA connected to a flame ionization detector (FID). This kind of instrument, is able to detect ions formed during combustion in a hydrogen flame. The generation of these ions is proportional to MeSA concentration in the sample stream. A calibration curve was initially created using samples with a well known pure MeSA concentration (0 mM, 0.2 mM, 0.5 mM, 1.0 mM, 2 mM).
NEB10β cells transformed with BBa_K1065102 were grown in M9 medium, induced with 5 mM arabinose and in some cases supplemented with salicylic acid. All the gas chromatography measures here reported were done in liquid phase, by injecting 1 ul of pre-filtered culture in the instrument.
Figure 1: induced sample produces MeSA. A culture of cells transformed with BBa_K1065102 was grown until O.D. 0.6 was reached. The culture was then splitted in 2 samples and one was induced with 5 mM arabinose. 2 mM salycilic acid was added to these samples. After about 4 h the samples were connected to the Gas Chromatograph. The induced sample (blue trace) shows the characteristic peak of methyl salicylate, as opposed to non induced cells (red trace).

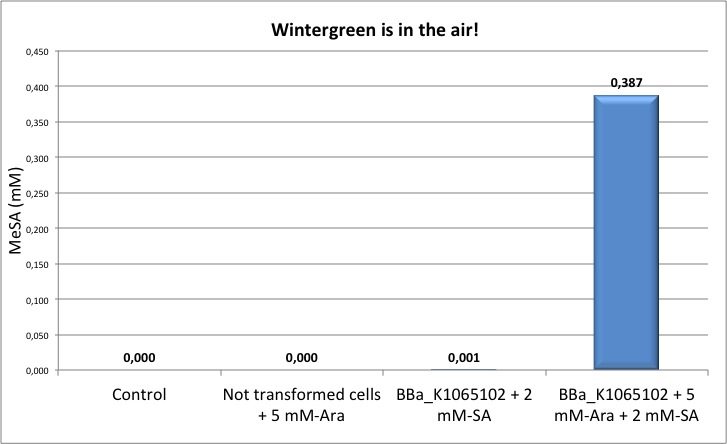
Figure 2: Left panel: calibration curve obtained with different concentrations of pure MeSA in ethanol. Right panel: Quantification of MeSA by GC-FID. NEB10β cells transformed with BBa_K1065102 supplemented with salycilic acid produce around 0.4 mM of MeSA. Non transformed cells and non induced cells did not produce any MeSA. Cells induced with arabinose and not supplemented with salycilic acid did not show any significant MeSA concentration (data not shown).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 3107
Illegal BamHI site found at 3856 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 389
Illegal NgoMIV site found at 793
Illegal NgoMIV site found at 918
Illegal NgoMIV site found at 929
Illegal NgoMIV site found at 1210
Illegal NgoMIV site found at 1678
Illegal AgeI site found at 2942 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 3397
Illegal SapI site found at 2924
| None |
