Promoters/Catalog/Anderson
Description
Members of the Anderson promoter collection are suitable for general protein expression in E. coli and likely other prokaryotes. The collection is known to cover a range of activities so by testing a few promoters it should be possible to find a promoter activity that suits your application. The promoters were recovered from a library screen by Chris Anderson.
Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. J23119 is the "consensus" promoter sequence and the strongest member of the family. The NheI and AvrII restriction sites present within these promoter parts make them a scaffold for further modification.
Obtaining Anderson promoter collection
The sequences of the Anderson promoters can be found via the table below. To obtain the physical DNA, we recommend two approaches -
Via de novo synthesis: Since the promoters are short sequences, they can be easily and cheaply ordered as two single-stranded complementary oligo's and annealed. See here for a tutorial on how to construct short parts via oligo annealing.
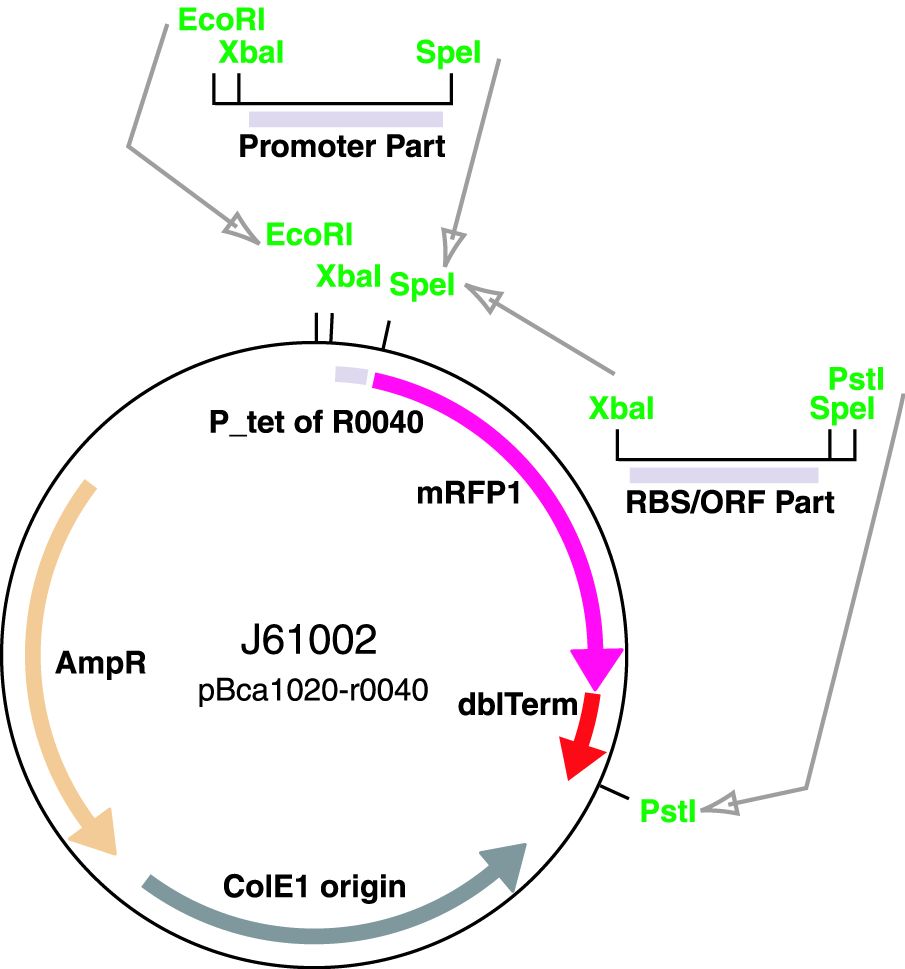
Via the Registry distribution: The promoters are included in the Registry distribution. All parts except BBa_J23119 are present in plasmid BBa_J61002. This places the RFP downstream of the promoter. Part BBa_J23119 is present in pSB1A2.
Anderson promoter collection
| Identifier | Sequencea | Predicted Strengthb | |
|---|---|---|---|
| BBa_J23119 | ttgacagctagctcagtcctaggtataatgctagc | n/a | |
| BBa_J23100 | ttgacggctagctcagtcctaggtacagtgctagc | 1 | |
| BBa_J23101 | tttacagctagctcagtcctaggtattatgctagc | 0.70 | |
| BBa_J23102 | ttgacagctagctcagtcctaggtactgtgctagc | 0.86 | |
| BBa_J23103 | ctgatagctagctcagtcctagggattatgctagc | 0.01 | |
| BBa_J23104 | ttgacagctagctcagtcctaggtattgtgctagc | 0.72 | |
| BBa_J23105 | tttacggctagctcagtcctaggtactatgctagc | 0.24 | |
| BBa_J23106 | tttacggctagctcagtcctaggtatagtgctagc | 0.47 | |
| BBa_J23107 | tttacggctagctcagccctaggtattatgctagc | 0.36 | |
| BBa_J23108 | ctgacagctagctcagtcctaggtataatgctagc | 0.51 | |
| BBa_J23109 | tttacagctagctcagtcctagggactgtgctagc | 0.04 | |
| BBa_J23110 | tttacggctagctcagtcctaggtacaatgctagc | 0.33 | |
| BBa_J23111 | ttgacggctagctcagtcctaggtatagtgctagc | 0.58 | |
| BBa_J23112 | ctgatagctagctcagtcctagggattatgctagc | 0.00 | |
| BBa_J23113 | ctgatggctagctcagtcctagggattatgctagc | 0.01 | |
| BBa_J23114 | tttatggctagctcagtcctaggtacaatgctagc | 0.10 | |
| BBa_J23115 | tttatagctagctcagcccttggtacaatgctagc | 0.15 | |
| BBa_J23116 | ttgacagctagctcagtcctagggactatgctagc | 0.16 | |
| BBa_J23117 | ttgacagctagctcagtcctagggattgtgctagc | 0.06 | |
| BBa_J23118 | ttgacggctagctcagtcctaggtattgtgctagc | 0.56 | |
aThe sequence of individual promoters are shown in black and red. The black nucleotides are part of the consensus promoter sequence (BBa_J23119) among all promoters while the red nucleotides highlight the differences between the individual promoters and the consensus sequence.
bThe relative strengths of these promoters were measured by Chris Anderson and the 2006 Berkeley iGEM team. These measurements are explained in greater detail in the Characterization section below.
Characterization
Measured strengths
Reported activities of the promoters are given as the relative fluorescence of these plasmids in strain TG1 grown in LB media to saturation. See part J61002 for details on their use.