Part:BBa_K914018
P1003** Kan resistant gene with 2 Amber Codon
The amber mutations avoid the expression of the P1003 gene even at usual concentration that prevent the expression of kanamycin resistance, although one mutation (BBa_K914009) permits some leakage in translation for this gene. This mutation is rescued with the tRNA amber suppressor supD (BBa_K914000). The idea here is to prevent the kanamycin gene resistance to be expressed in case horizontal gene transfer.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
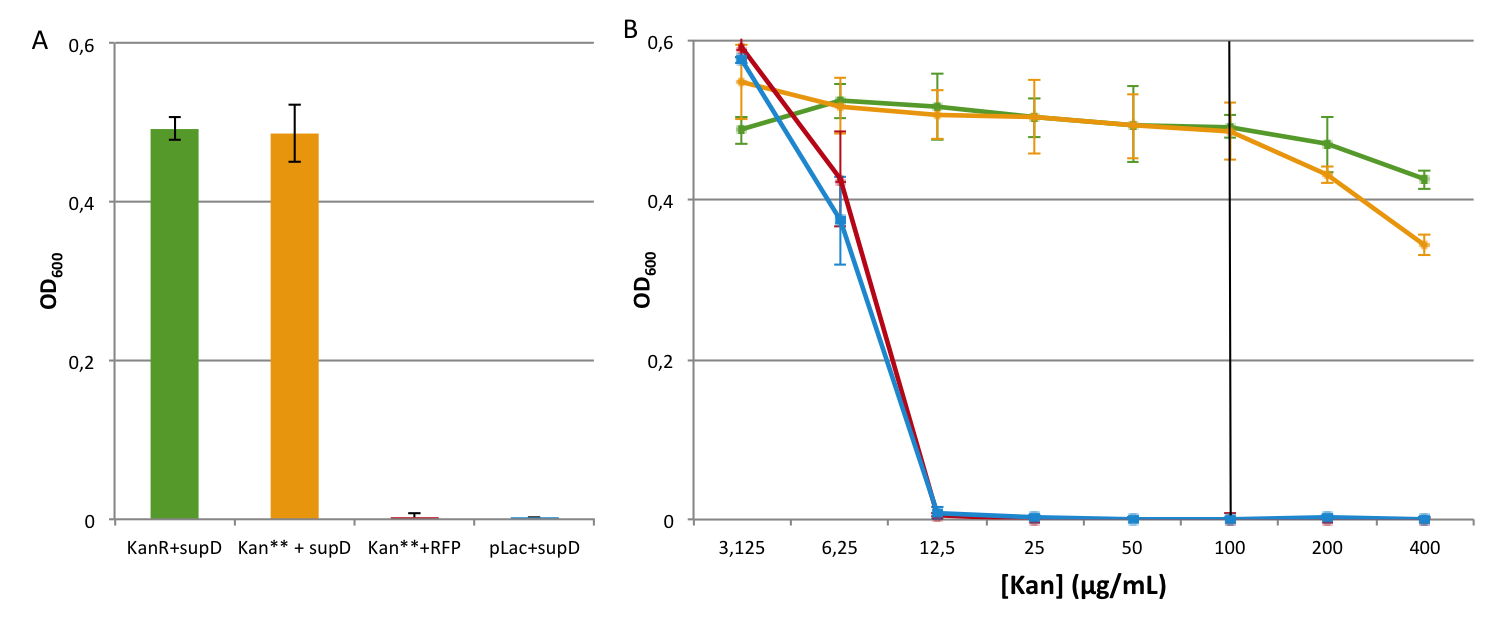
The positive and negative controls behave correctly, and similarly to the previous experiment (see K914009), it is also the case for the complementation of Kan** with supD. That means that two mutation are not a problem for the cell. Moreover, we notice that there is no more leakiness in the system at usual concentration, even at low concentration. Actually the double kanamycin resistant gene with two mutation behave exactly like the negative control (Plac + supD) which means that semantic containment is working well.
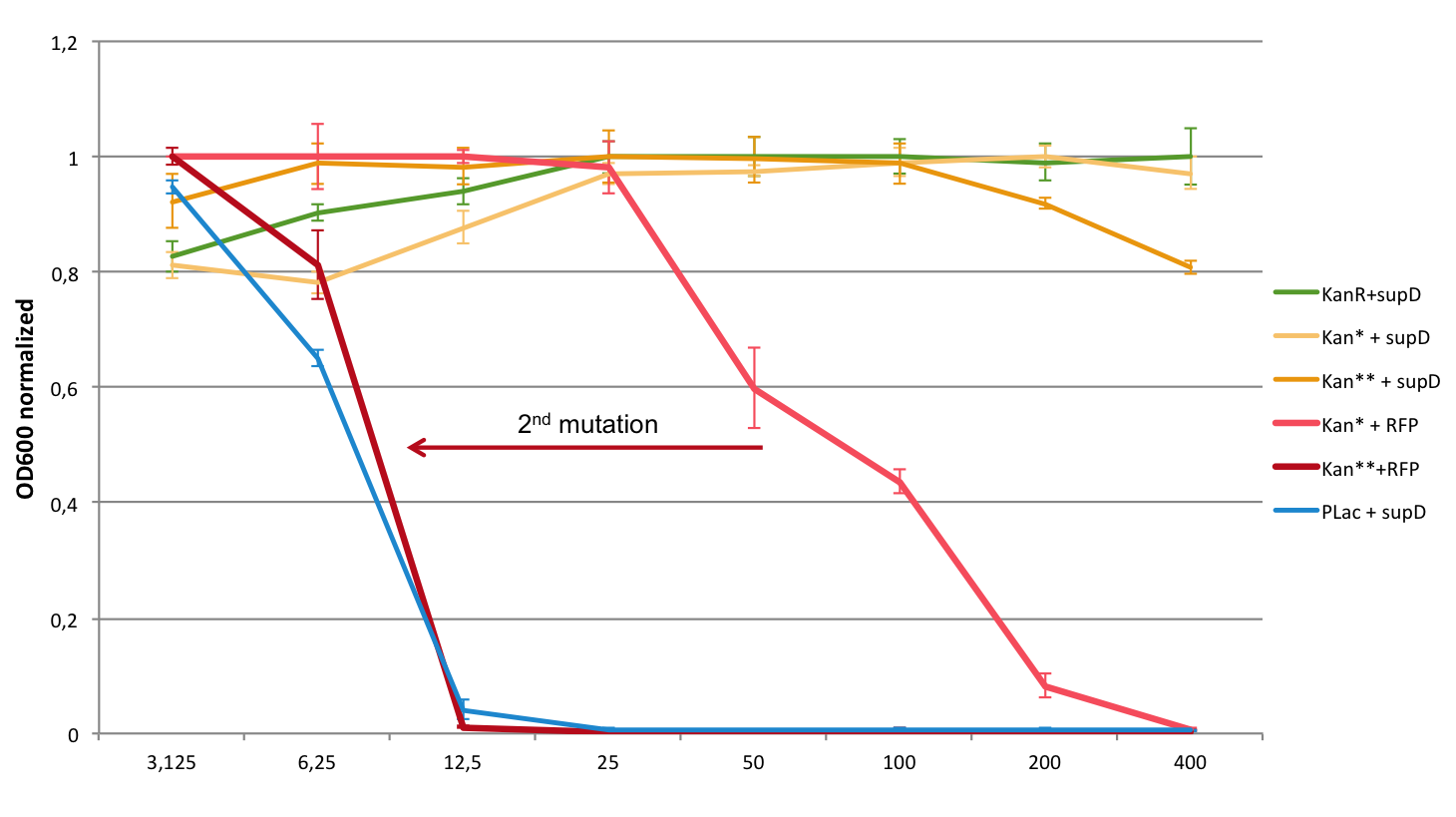
Comparison of K914009 (Kan*) and K914018 (Kan**)
Here is a graph that gather both results, to highlight the effect of the second mutation. The results are normalized intra group (experiment with the one-mutated gene, and with the two-mutated gene) in order to have the same scale to compare them. We took the average of the positive (KanR+supD) and the negative (Plac+supD) controls.
Functional Parameters
//cds/selectionmarker/antibioticresistance
| None |