Part:BBa_E0040
green fluorescent protein derived from jellyfish Aequeora victoria wild-type GFP (SwissProt: P42212
GFP (mut3b) [note that this part does not have a barcode]
Usage and Biology
Untagged version of gfp from Repressilator reporter. See the design page for more source information.
The original citation for GFPmut3b is as follows:
Cormack, B.P., Valdivia, R.H., and S. Falkow. FACS-optimized mutants of green fluorescent protein (GFP). Gene 173: 33-38 (1996).
Here's the link: http://www.sciencedirect.com/science/article/pii/0378111995006850
Fluorescence wavelengths
Cormack et al.Cormack report the following excitation and emission data for GFPmut3 -
- Excitation max - 501nm
- Emission max - 511nm
Latency
Cormack et al.Cormack report detectable fluorescence within 8 mins. Please add maturation time data for E0040 here.
References
<biblio>
- Cormack pmid=10659856
</biblio>
Part Characteristics in Cell-Free Chassis
| Parameter | Value and Description |
|---|---|
| Calibration | A conversion factor of 79.429 from Au to concentraion in nM |
| Half-life | 33 hours in the cell-free chassis, with a degradation constant of 0.0210 (in hours) |
Purificaton
GFPmut3b can be purified for calibration after the addition of a his-tag. The detailed [http://2007.igem.org/Imperial/Wet_Lab/Protocols/Prot1.6 protocols]and [http://2007.igem.org/Imperial/Wet_Lab/Results/Res1.6 results]for the purification can be found.
Calibration
The fluorescence of purified GFPmut3B was calibrated in the cell-free chassis. The derived [http://2007.igem.org/Imperial/Wet_Lab/Results/Res1.3 calilbration curve]allows the determination of the concentration of GFPmut3b in the cell-free chassis. [http://2007.igem.org/Imperial/Wet_Lab/Protocols/Prot1.3 Detailed protocols]for generating the calibration curve are available. Other calibration curves for are also available on the results page.
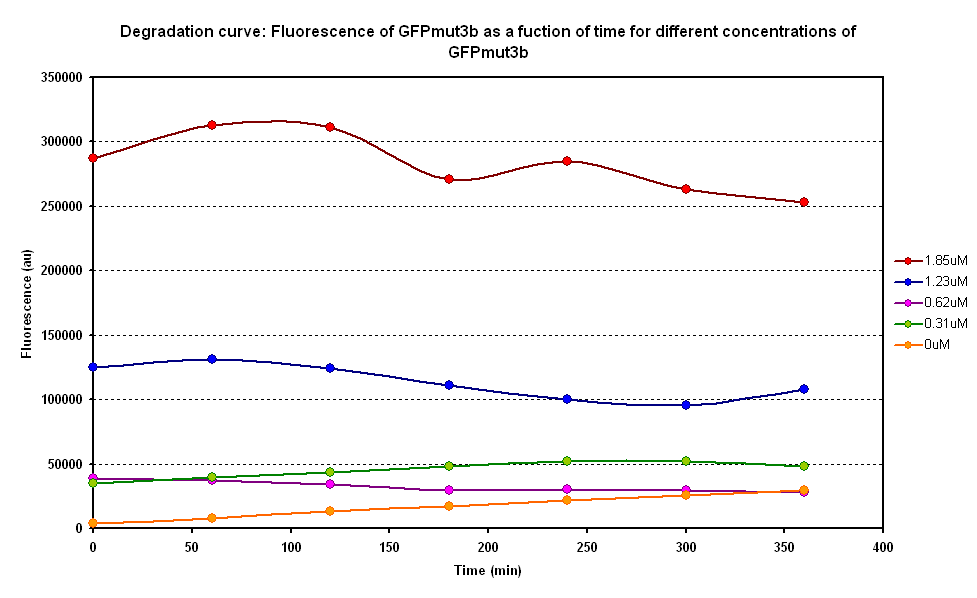
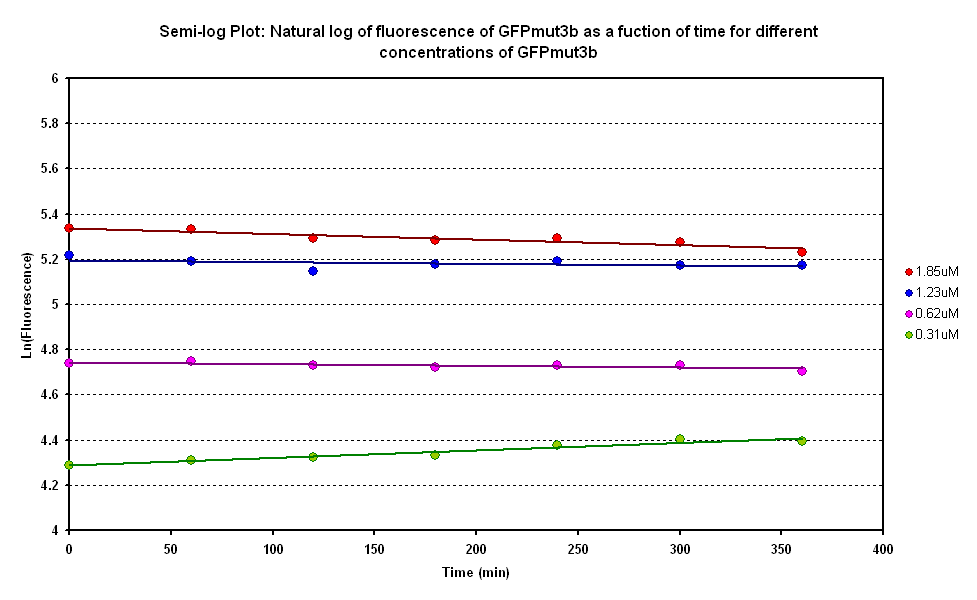
Degradation
The degradation of GFPmut3B in the cell-free chassis was also characterized. Purified GFPmut3B was allowed to degrade in the cell-free chassis and the fluorescence was measured over time. [http://2007.igem.org/Imperial/Wet_Lab/Protocols/Prot1.4 Detailed protocols]and [http://2007.igem.org/Imperial/Wet_Lab/Results/Res1.4 results]are attached.
From the semi-log plot, the degradation constant (in minutes) was derived to be 0.0003501, which is equivalent to GFPmut3b having a half-life of 33 hours in the cell-free chassis.
Improvements
Yeast- and FACS optimized GFP: BBa_K194001 Yeast- and FACS optimized, fast degradable GFP: BBa_K194002
| Protein data table for BioBrick BBa_E0040 automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 10: (underlined part encodes the protein) ATGCGTAAA ... CTATACAAATAATAA ORF from nucleotide position 1 to 714 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
Alignments (obtained from PredictProtein.org)
| ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Localization (reliability in brackets)
| Gene Ontology (reliability in brackets)
| |||||||||||||||||||||||||||||||||||||||||||||
Predicted features:
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
//chassis/prokaryote/bsubtilis
//chassis/prokaryote/ecoli
//function/reporter/fluorescence
| color | Green |
| direction | Forward |
| emission | |
| emit | 511 |
| excitation | |
| excite | 501 |
| kegg | |
| lum | |
| protein | GFPmut3b |
| swisspro | |
| tag | None |

 1 Registry Star
1 Registry Star