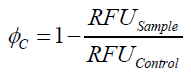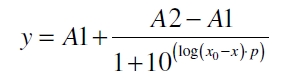Part:BBa_K525305
Fusion Protein of S-Layer SgsE and mCitrine
Fusion protein of S-layer SgsE and mCitrine
S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self assembly-domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to eachother (determined by the S-layer geometry) it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr et al., 2007]).
Usage and Biology
S-layer proteins can be used as scaffold for nanobiotechnological applications and devices by e.g. fusing the S-layer's self-assembly domain to other functional protein domains. It is possible to coat surfaces and liposomes with S-layers. A big advantage of S-layers: after expressing in E. coli and purification, the nanobiotechnological system is cell-free. This enhances the biological security of a device.
This fluorescent S-layer fusion protein is used to characterize purification methods and the S-layer's ability to self-assemble on surfaces. It is also possible to use the characteristic of mCitrine as a pH indicator ([http://pubs.acs.org/doi/abs/10.1021/bm901071b Kainz et al., 2010]).
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression (E. coli) | Localisation | Inclusion body |
| Compatibility | E. coli KRX and BL21(DE3) | |
| Inductor for expression | T7 polymerase + IPTG or lactose | |
| Purification | Molecular weight | 110.2 kDa |
| Theoretical pI | 5.74 | |
| Excitation / emission | 515 / 529 nm | |
| Immobilization behaviour | Saturation protein / bead ratio | 5 - 7 * 10-4 |
| Immobilization time | 5 h |
Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 167
Illegal BglII site found at 1022 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 76
Illegal AgeI site found at 3121 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1657
Expression in E. coli
Purification of SgsE fusion protein
Immobilization behaviour
Kinetic of refolding and immobilization to silicium dioxide beads
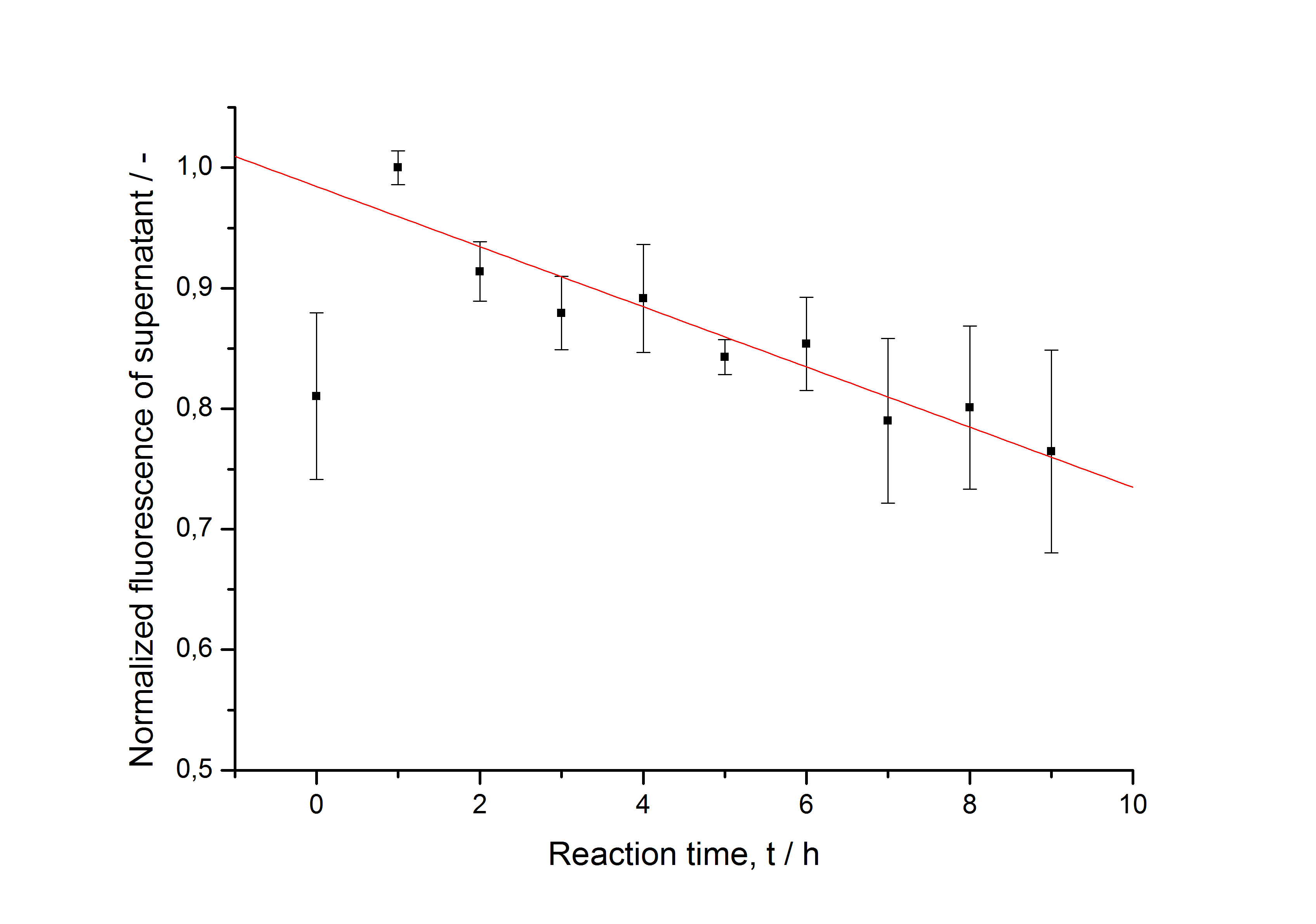
After purification, the SgsE fusion proteins are solved in 6 M urea so they have to be refolded and reassembled during immobilization. This kinetic is determined by starting several parallel immobilization experiments. Every hour, three experiments including beads and HBSS buffer instead of beads are centrifuged, the supernatant is collected and stored on ice. After washing the beads twice with ddH2O, they are stored in ddH2O. The fluorescence is measured in a Tecan reader. The data is normalized to the maximal value of fluorescence and plotted against the reaction time. The development of the fluorescence in the control without beads is shown in fig. B and on the beads in fig. C:
The fluorescence in the negative control rises in the first hour and then constantly decreases. This indicates, that the refolding of the fluorescent protein needs about one hour. After this hour the decrease shows that the protein is not very stable in the solution. Fused to the S-layer and immobilized on beads, this decrease is not observed, indicating that the immobilization enhances the stability of the fluorescent protein in solution. The maximal fluorescence on the beads is reached after 5 hours of incubation.
The data in fig. B is fitted (without including the t0 sample) with a linear function. The data in fig. C is fitted with the following hill function
with the Hill coefficient n and a constant k.
Optimal bead to protein ratio for immobilization
To determine the optimal ratio of silica beads to protein for immobilization, the degree of clearance ϕC in the supernatant is calculated and plotted against the concentration of silica beads used in the accordant immobilization experiment (compare fig. A):
The data was collected in three indipendent experiments. The fluorescence of the samples was measured in the supernatant of the immobilization experiment after centrifuging the silica beads. The fluorescence of the control was measured in a sample which was treated exactly like the others but no silica beads were added. 100 µg protein was used for one immobilization experiment. The data was fitted with a sigmoidal dose-response function of the form
with the Hill coefficient p, the bottom asymptote A1, the top asymptote A2 and the switch point log(x0) (R² = 0.874).
The fit indicates that a good silica concentration for 100 µg of protein is 150 - 200 mg mL-1. This set-up leads to saturated beads with low waste of protein. So a good protein / bead ratio to work with is 5 - 7 * 10-4.

Methods
References
Kainz B, Steiner K, Möller M, Pum D, Schäffer C, Sleytr UB, Toca-Herrera JL (2010) Absorption, Steady-State Fluorescence, Fluorescence Lifetime, and 2D Self-Assembly Properties of Engineered Fluorescent S-Layer Fusion Proteins of Geobacillus stearothermophilus NRS 2004/3a, [http://pubs.acs.org/doi/abs/10.1021/bm901071b Biomacromolecules 11(1):207-214].
Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM (2007) S-layers as a tool kit for nanobiotechnological applications, [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full FEMS Microbiol Lett 267(2):131-144].
//function/reporter/fluorescence
//proteindomain/internal
| biology | S-Layer |
| chassis | E. coli |
| emission | 529 nm |
| excitation | 515 nm |
| origin | Geobacillus stearothermophilus |