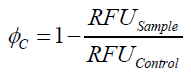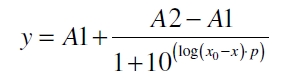Part:BBa_K525305
Fusion Protein of S-Layer SgsE and mCitrine
Fusion protein of S-layer SgsE and mCitrine
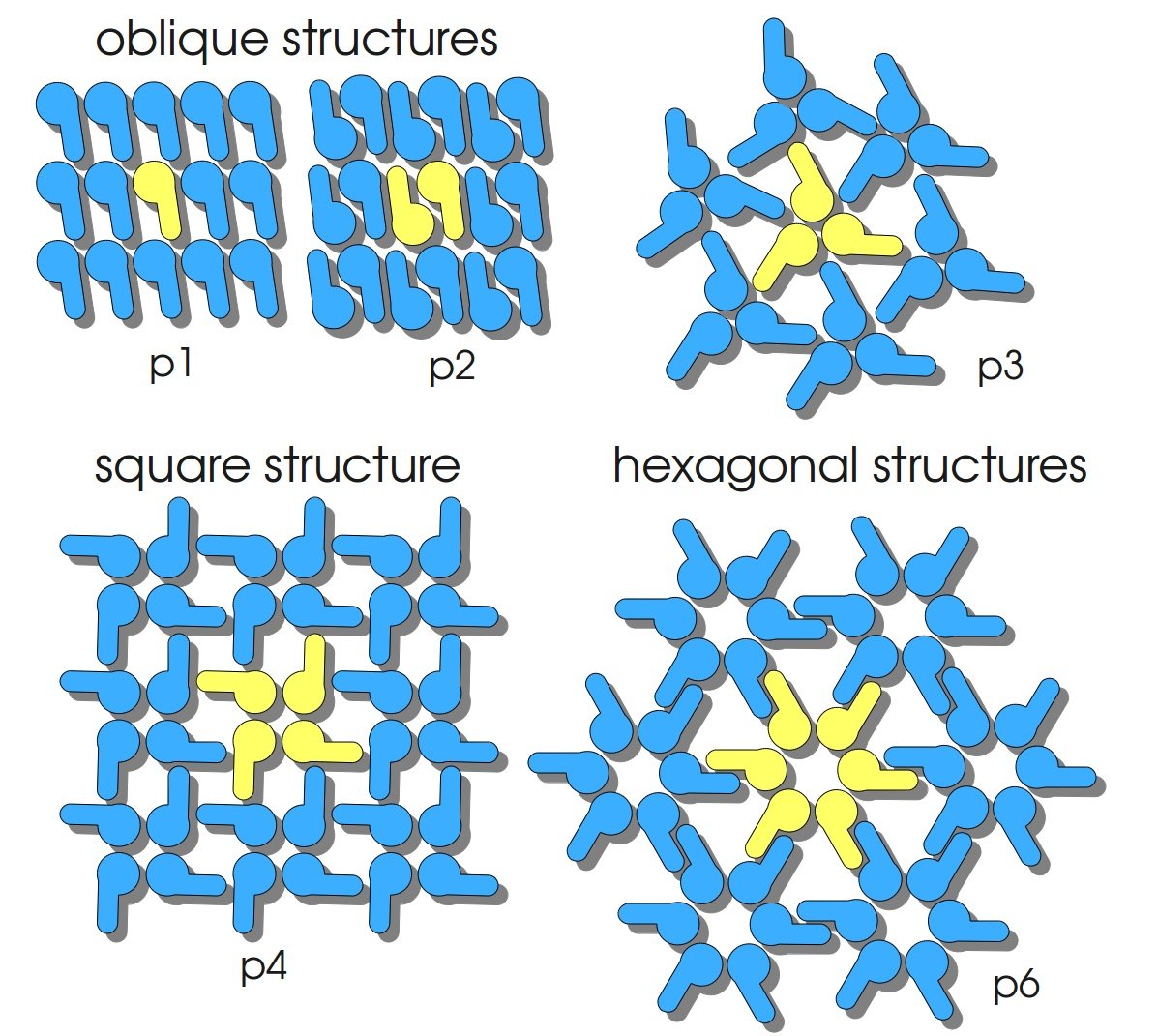
S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self assembly-domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to eachother (determined by the S-layer geometry) it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr et al., 2007]).
Usage and Biology
S-layer proteins can be used as scaffold for nanobiotechnological applications and devices by e.g. fusing the S-layer's self-assembly domain to other functional protein domains. It is possible to coat surfaces and liposomes with S-layers. A big advantage of S-layers: after expressing in E. coli and purification, the nanobiotechnological system is cell-free. This enhances the biological security of a device.
This fluorescent S-layer fusion protein is used to characterize purification methods and the S-layer's ability to self-assemble on surfaces. It is also possible to use the characteristic of mCitrine as a pH indicator ([http://pubs.acs.org/doi/abs/10.1021/bm901071b Kainz et al., 2010]).
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression (E. coli) | Localisation | Inclusion body |
| Compatibility | E. coli KRX and BL21(DE3) | |
| Induction of expression | expression of T7 polymerase + IPTG or lactose | |
| Inhibition of expression | glucose | |
| Specific growth rate (un-/induced) | 0.139 h-1 / 0.071 h-1 | |
| Doubling time (un-/induced) | 4.98 h / 9.78 h | |
| Purification | Molecular weight | 110.2 kDa |
| Theoretical pI | 5.74 | |
| Excitation / emission | 515 / 529 nm | |
| Immobilization behaviour | Saturation protein / bead ratio | 5 - 7 * 10-4 |
| Immobilization time | 4 h | |
| lattice structure | oblique (p2) |
Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 167
Illegal BglII site found at 1022 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 76
Illegal AgeI site found at 3121 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1657
Expression in E. coli
The SgsE gene under the control of a T7 / lac promoter (BBa_K525303) was fused to mCitrine (BBa_J18931) using Freiburg BioBrick assembly for characterization experiments.
The SgsE|mCitrine fusion protein was overexpressed in E. coli KRX after induction of T7 polymerase by supplementation of 0.1 % L-rhamnose and 1 mM IPTG using the autoinduction protocol by Promega.

Purification of SgsE fusion protein
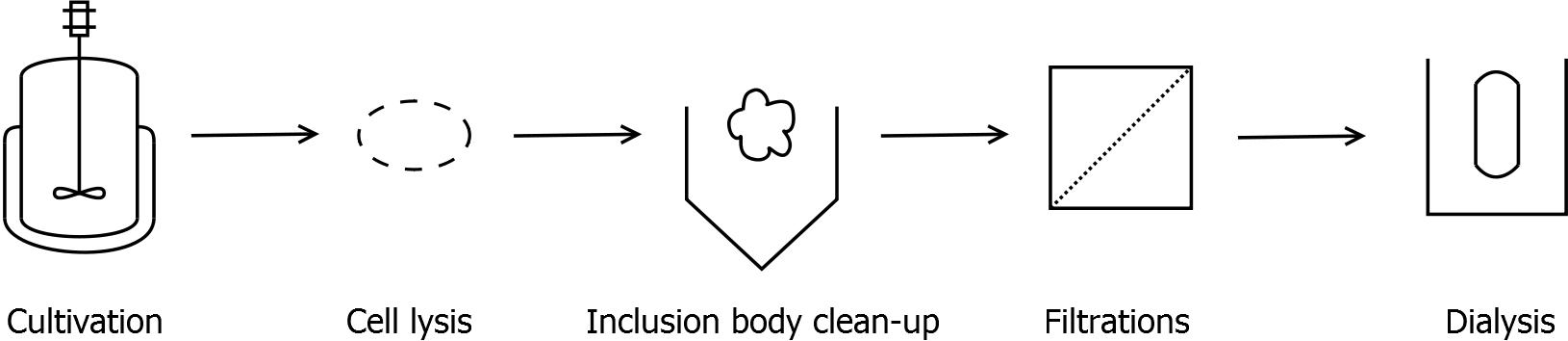
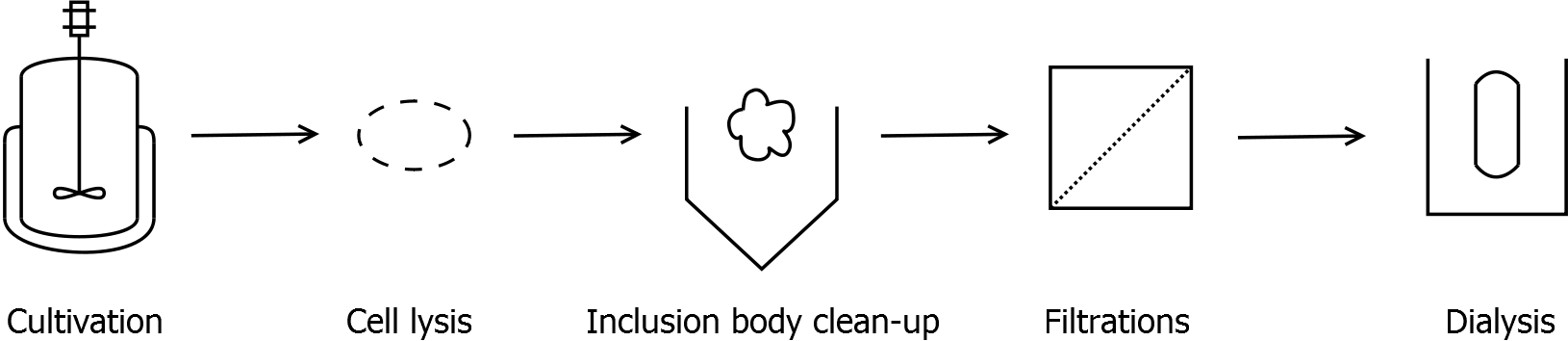
Strategy without His-tag
Scheme of purification strategy for SgsE (fusion) proteins without His-tag:
As observed in the analysis of the cultivations with expression of SgsE | mCitrine fusion proteins, these proteins form inclusion bodies in E. coli. Inclusion bodies have the advantage that they are relatively easy to clean-up and are resistant to proteases. The first purification step is to isolate and solubilize the inclusion bodies. This step is followed by two filtrations (300 kDa UF and 100 kDa DF/UF) to further concentrate and purify the S-layer proteins. After the filtrations, the remaining protein solution is dialyzed against ddH2O for 18 h at 4 °C in the dark. The dialysis leads to a precipitation of the water-insoluble proteins. After centrifugation of the dialysate, the water-soluble S-layer monomers remain in the supernatant and can be used for recrystallization experiments.
The fluorescence of the collected fractions of this purification strategy is shown in the following figure 3:

A huge amount of protein is lost during the purification, especially after the centrifugation steps. The fluorescence in the urea containing fractions is lowered due to denaturation of the fluorescent protein. Some fluorescence could be regenerated by the recrystallization in HBSS. This purification strategy is very simple and can be carried out by nearly everyone in any lab, being the first step to enable do it yourself nanobiotechnology.
Strategy with His-tag
Scheme of purification strategy for SgsE (fusion) proteins with His-tag:
By fusing the SgsE | mCitrine with a C-terminal His-6-tag the S-layer protein could be simply purified by using a denaturating His-tag affinity chromatography. This purification strategy has the advantage that no time-consuming and complex inclusion body purification and filtration is necessary to decrease the amount of native E. coli proteins. Additional to the simplification a higher purity of the S-layer protein could be reached.
This purification was performed using the SgsE fusion protein containing a N-terminal mCitrine,for identification and a C-terminal His-6-tag. The SDS-PAGE gel (Fig. 4) and the fluorescence (Fig. 5) in the collected fractions showed that the majority of fusion protein was eluated with a imidazole concentration of approx. 50 mM. There was also fluorescence measurable in the flowthrough and wash fractions. This indicates that the used 1 mL HisTrap FF crude (GE Healthcare) was overloaded or the protein was bound weakly at to the affinity matrix. The resulting purity in the elution fraction, the saving of time and the simplicity makes this procedure the prefered purification method.


Immobilization behaviour
After purification, solutions of monomeric SgsE S-layer proteins can be recrystallized and immobilized on silicon dioxide beads in HBSS (Hank's buffered saline solution). After the recrystallization procedure the beads are washed with and stored in ddH2O at 4 °C in the dark. The fluorescence of the collected fractions of a recrystallization experiment with BBa_K525305 are shown in Fig. 6. 100 mg beads were coated with 100 µg of protein. The figure shows, that not all of the protein is immobilized on the beads (supernatant fraction) but the immobilization is quite stable (very low fluorescence in the wash). After the immobilization, the beads display a high fluorescence indicating the binding of the SgsE | mCitrine fusion protein.

Optimal bead to protein ratio for immobilization
To determine the optimal ratio of silica beads to protein for immobilization, the degree of clearance ϕC in the supernatant is calculated and plotted against the concentration of silica beads used in the accordant immobilization experiment (compare Fig. 7):
The data was collected in three independent experiments. The fluorescence of the samples was measured in the supernatant of the immobilization experiment after centrifugation of the silica beads. The fluorescence of the control was measured in a sample which was treated exactly like the others but no silica beads were added. 100 µg protein was used for one immobilization experiment. The data was fitted with a sigmoidal dose-response function of the form.
with the Hill coefficient p, the bottom asymptote A1, the top asymptote A2 and the switch point log(x0) (R² = 0.874).
The fit indicates that a good silica concentration for 100 µg of protein is 150 - 200 mg mL-1. This set-up leads to saturated beads with low waste of protein. So a good protein / bead ratio to work with is 5 - 7 * 10-4.

Methods
Expression of S-layer genes in E. coli in shaking flasks
- Chassis: Promega's [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ E. coli KRX]
- Medium: LB medium supplemented with 20 mg L-1 chloramphenicol
- For autoinduction: Cultivations in LB-medium were supplemented with 0.1 % L-rhamnose and 1 mM IPTG as inducer and 0.05 % glucose
- cultivation: 37 °C with 120 rpm in shaking flasks
Production of SgsE
Cultivation
- Bioreactor: [http://www.bioengineering-inc.com/standard-reactors.php?id=2.1 Bioengineering NLF22 7 L] with Bioengineering DCU
- Medium: [http://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#HSG_medium HSG medium] with 20 mg L-1 chloramphenicol
- Culture volume: 4 L
- Inoculation OD600: 0.2
- DO: 40 % airsaturation (controlled with stirrer cascade starting with 200 rpm)
- pH: 7.0 (controlled with 20 % phosphoric acid and 2 M NaOH)
- Induction after 4 h cultivation time with 2 % rhamnose and 0.1 mM IPTG (in culture medium)
- Harvest after 13 h
Cell lysis
- For big amounts
- Centrifuge down the cells (10000 g, 30 min, 4 °C)
- Resuspend pellet in enzyme buffer (constructs without His-tag) or binding buffer (constructs with His-tag)
- Cell lysis with high-pressure homogenizer (800 bar, 3 cycles at 4 °C)
- Centrifuge down the lysate (10000 g, 60 min, 4 °C)
- For small amounts
- Harvest cells by centrifugation at 10000 g for 10 min at 4 °C
- Resuspend the pellet in 5 mL enzyme buffer (constructs without His-tag) or binding buffer containing 10 mM imidazole (constructs with His-tag) for each gram of cell paste
- Sonification on ice for approx. 5 min with Sonifier 450 by [http://www.gehealthcare.com/ Branson]
- Centrifuge at 10000 g for 30 min at 4 °C
Purification for constructs with His-tag
- Denaturating immobilized Metal Affinity Chromatography (IMAC)
- Column: 1 mL HisTrap FF crude by [http://www.gehealthcare.com/ GE Healthcare] (Ni-NTA)
- Wash column with 5 - 10 mL of deionized water
- Equilibrate column with 5 - 10 mL of binding buffer
- Load sample onto the column
- Wash with 10 mL binding buffer
- Elute with elution buffer with increasing imidazole concentrations
- Collect the eluate fractions, the purified protein is most likely in the fractions containing 50 - 75 mM imidazole
- Re-equilibrate the column with binding buffer
Purification for constructs without His-tag
- Inclusion body clean-up
- wash pellet from cell lysis with water twice
- after washing the pellet: incubate the pellet in [http://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Denaturation_buffer_for_inclusion_bodies denaturation buffer] for 60 min, 4 °C with vertical rotator
- final concentration in denaturation buffer: 0.5 mg wet biomass per mL
- centrifuge (60 min, >17000 g, 4 °C)
- in general with all centrifugations during this clean-up: the higher the speed, the better the result
- collect supernatant and incubate the pellet again in denaturation buffer (60 min, 4 °C, vertical rotator)
- centrifuge (60 min, >17000 g, 4 °C)
- collect supernatant and discard pellet

- Filtration
- Arrange the filtration module as shown on the right side.
- Collect permeate of cross flow filtration with 300 kDa membrane of sample before ultrafiltration
- This step is for removing cell debris
- Diafiltrate with 100 kDa membrane against [http://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Denaturation_buffer_for_inclusion_bodies denaturation buffer]
- constantly delute permeate with the buffer, keeping the permeate volume as low as possible
- Used membranes: [http://www.millipore.com/catalogue/module/C7493 Milipore Pellicon XL 50] or XL 100 membranes
- 50, 100 or 300 kDa cut-off
- 50 cm2 filtration area
- tangential flow filter
- Hydrophilic polyvinylidene fluoride membrane
- Used pump: SciLog TANDEM 1081 peristaltic pump
- flow rate during filtration: 40 mL min-1
- Dialysis
- Fill retentate from DF/UF in dialysis tube ([http://www.carl-roth.de/ Roth], cellulose, 10 kDa cut-off)
- Dialyse against ddH2O for 18 h at 4 °C in the dark
- After dialysis: centrifuge down the precipitation (45 min, 17000 g, 4 °C) and collect the supernatant
- Measure protein concentration in supernatant, dilute to 1 mg mL-1 with ddH2O and store at 4 °C in the dark
Measuring of mCitrine
- Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination
- Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark
- To measure the samples thaw at room temperature and fill 200 µL of each sample in one well of a black, flat bottom 96 well microtiter plate (perform at least a repeat determination)
- Measure the fluorescence in a platereader (we used a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]) with following settings:
- 20 sec orbital shaking (1 mm amplitude with a frequency of 87.6 rpm)
- Measurement mode: Top
- Excitation: 488 nm
- Emission: 529 nm
- Number of reads: 25
- Manual gain: 100
- Integration time: 20 µs
References
Kainz B, Steiner K, Möller M, Pum D, Schäffer C, Sleytr UB, Toca-Herrera JL (2010) Absorption, Steady-State Fluorescence, Fluorescence Lifetime, and 2D Self-Assembly Properties of Engineered Fluorescent S-Layer Fusion Proteins of Geobacillus stearothermophilus NRS 2004/3a, [http://pubs.acs.org/doi/abs/10.1021/bm901071b Biomacromolecules 11(1):207-214].
Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM (2007) S-layers as a tool kit for nanobiotechnological applications, [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full FEMS Microbiol Lett 267(2):131-144].
//function/reporter/fluorescence
//proteindomain/internal
| biology | S-Layer |
| chassis | E. coli |
| emission | 529 nm |
| excitation | 515 nm |
| origin | Geobacillus stearothermophilus |