Part:BBa_K1162006
LL-37 antimicrobial peptide from Humans (Homo sapiens)
LL-37 is an antimicrobial peptide (AMP) from the cathelicidin family of antimicrobial peptides, isolated from the Human (Homo sapiens). It is a 37-residue helical peptide found throughout the human body, exhibiting a broad spectrum of antimicrobial activity as well as a variety of immunomodulating effects (Dürr, Ulrich H.N., et. al 2006). It has the distinction of being the first and only cathelicidin member discovered from humans, and is extremely well studied and characterized for this reason. The peptide is produced via epithelial cells as well as leukocytes in places such as the skin, gastrointestinal tract, and the respiratory tract. This AMP is is cleaved from a larger protein, hCAP-18, in a series of modifications to yield its active form (Sorensen, O.E, et. al 2001).
Design
This part was codon optimized for E. coli using the Life Technologies GeneArt ® software program. By design, this part is Assembly Standard #23 (Silver Fusion) and contains a Methionine(atg) residue at the start of the coding region. Furthermore, this LL-37 sequence does not have any stop codons, which allows for composite BioBrick construction.
Improvement on Existing Parts (as of September 2013)
BBa_K245114 (Slovenia iGEM team 2009) - This part lacks a start codon, which would require additional design to be expressed individually. The BBa_K1162006 part improves upon this one by adding a Met(atg) residue at the beginning of the coding region. It is unclear if Slovenia's part has been codon optimized for E. coli, but our new part has been designed with this in mind by Life Technologies (see design).
BBa_K875009 (Trieste iGEM team 2012) - This part, BBa_K875009, uses the Assembly Standard #10 for both the prefix and suffix and therefore cannot be used for protein fusions, such as in cases of secretion, protein purification, or fusion proteins. This part also has a stop codon which further limits what can be added to the C-terminal end (purification tag/secretion tag). The BBa_K1162006 improves upon this part by using Assembly Standard #23 (Silver Fusion) and by removing the stop codon at the end of the coding region. Additionally, the BBa_K1162006 part was codon optimized by Life Technologies (see design).
Experience

Figure 1. The image above of protein purification fractions demonstrates that there is expression of LL-37 and WAM-1 in E. coli through GFP fluorescence (GFP is at the C-terminal end). Unfortunately, there were issues with binding to the nickel affinity column, which will be troubleshooted in future work.

Figure 2. This image is an SDS-PAGE gel of the protein products from a 10L fermentation run for the LL37-spider silk generating construct using E. coli as the host organism. In the elution fraction of the gel image below, we can see a protein band at approximately 55-60 kDa, the size of the LL37-spider silk protein. From this study we have demonstrated that antimicrobial spider silk can be produced in E. coli and scaled-up. Since we also have additional AMP-spider silk constructs (see [http://2013.igem.org/Team:Utah_State/Parts BioBricks] page for a complete list), the next step would be to manufacture more of these using a similar approach to the LL37-spider silk. In future work, optimization of LL37-spider silk production will also take place.
References
Dürr, Ulrich H.N. Sudheendra, U.S. and Ramamoorthy, Ayyalusamy, LL-37, the only human member of the cathelicidin family of antimicrobial peptides (Review). Biochimica et Biophysica Acta 1758 (2006) 1408–1425.
O.E. Sorensen, P. Follin, A.H. Johnsen, J. Calafat, G.S. Tjabringa, P.S. Hiemstra, N. Borregaard, Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3, Blood 97 (2001) 3951–3959.
Characterization: Jilin_China 2019
Group:
Jilin_China 2019Authors:
Chang YangSummary:
On the basis of BBa_K1162006, we added the termination codon and characterized it.Documention:
1. Characterization
1.1 Validation of LL-37 construction
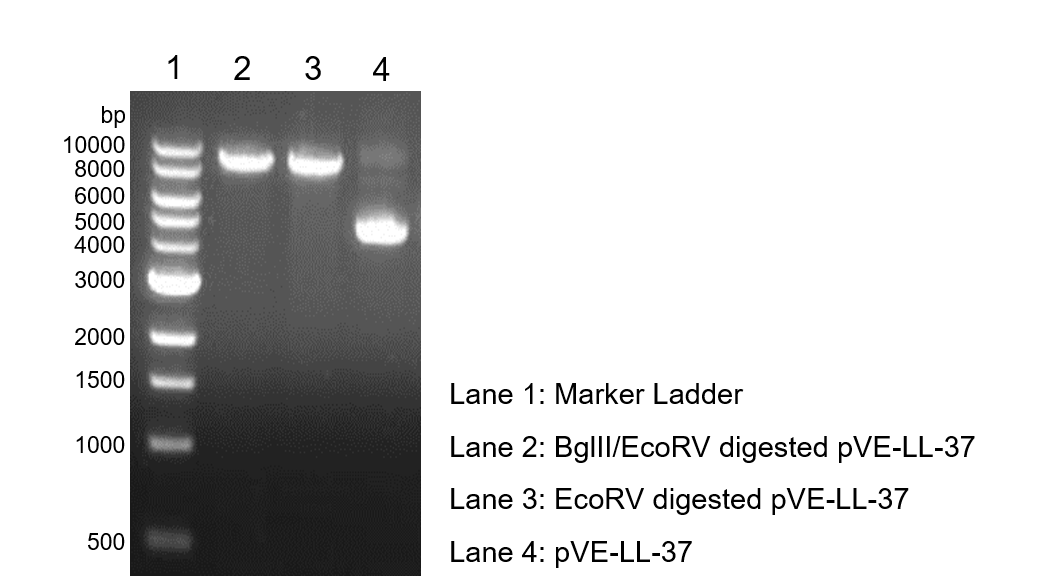
To verify the construction of pVE-LL-37 which we generated, the digestion by BglII/EcoRV was performed by a standard protocol followed by agarose gel electrophoresis (Figure 1).
Figure 1. Digestion and electrophoresis of pVE-LL-37.
1.2 Expression of pVE-LL-37
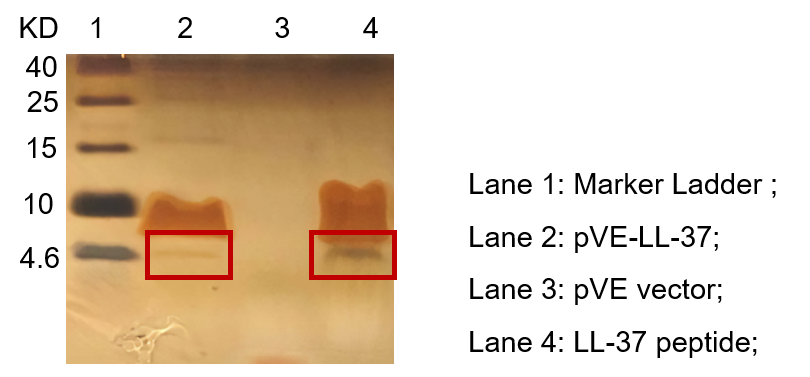
To detect the expression of pVE-LL-37 in E. coli, the constructs were transformed into BL21. Compared to the mock, there was a small size band shown in pVE-LL-37 (Figure 2).
Figure 2. Expression of pVE-LL-37. The bacteria were collected and ultrasonicated. The lysate was centrifuged and supernate was electrophoresed on the Tricine-SDS-PAGE gel, followed by Fast silver staining. The LL-37 peptide was synthesized as positive control.
1.3 Candidacidal effect of LL-37
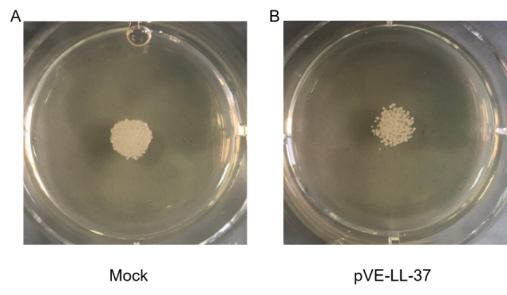
To assess the candidacidal activity of LL-37, the spot assay was performed. As shown in Figure 3, C. albicans viability after LL-37 treatment was visually compared with that of control cells (no LL-37 treatment). We found that cell viability was more sensitive to LL-37.
Figure 3. Candidacidal activity of LL-37. C. albicans were collected and suspensions were resuspended to 105 cells/mL. 10 µL C. albicans suspension and 20 µL sterile BL21-transformed bacteria were gently mixed. After a 10 min incubation, the mixture was spotted onto the SDA agar plates. Cell viability was detected after incubation at 30℃ for 18 hrs.
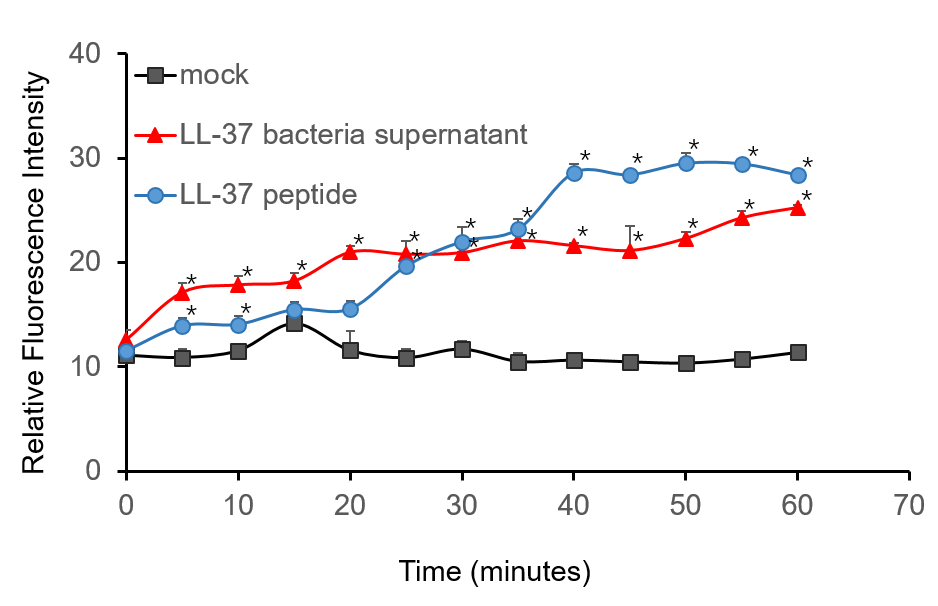
To further validate the candidacidal effect of LL-37, C. albicans suspensions were incubated with it. After incubation, LL-37 caused an immediate increase in PI fluorescence indicates that LL-37 can kill C. albicans (Figure 4).
Figure 4. Candidacidal activity of LL-37. 1.5 µM PI (propidiumiodide) was added into C. albicans suspensions (106 cells/mL, 1 mL) and serially BL21-transformed bacteria supernatant (2 mL) mixture. The whole system was incubated at 30 ℃. During the 1 h incubation, the fluorescence was measured every 5 min at the excitation wavelength λexc 544 nm and emission wavelength λem 620 nm. The experiment was performed three times in triplicate. *, P < 0.05 from control using Student’s t test.
3. Conclusion
We successfully characterized LL-37 and demonstrated its ability to kill C. albicans from multiple angles.
HZAU-China 2021
Design
In order to fit the project and guide the experiment, our research decided to depict the characterization of LL-37 protein- an antimicrobial peptide.
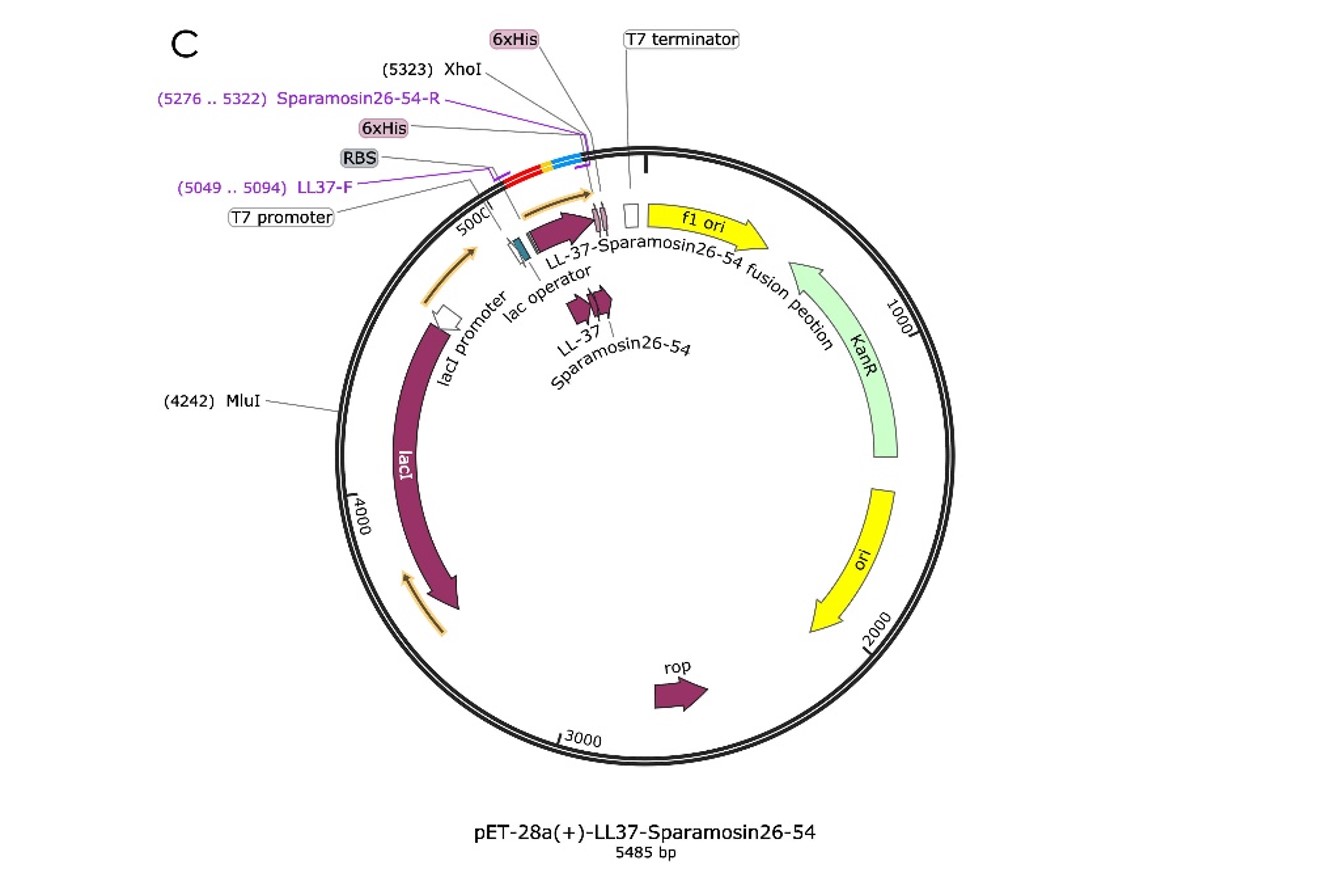
The plasmid we designed consists of T7 promoter, lac operator, RBS, LL-37 coding sequence, His tag and T7 terminator, which are arranged in an order on a pET28 backbone. We aim to induce the transcription of the downstream LL-37 by adding the IPTG to initiate the expression. The protein will then be purified and antibacterial activity was tested by culturing the Salmonella enterica subsp. enterica (ex Kauffmann and Edwards) Le Minor and Popoff serovar Typhimurium with our purified LL-37 and measuring the OD600 to reflect the bacteria growing condition.
Expectation
OD600 itself or the growing rate is expected to descend, which represent that the growth of bacteria is inhabited.
Materials and Method
1. Plasmids Construction: LL-37 is constructed using primers by overlap extension PCR. The constructed fragment is ligated to the target site in pET28 by PCR and homologous recombination. The correct construction is confirmed by sequencing.
2. Expression and Purification
1) Plasmid pET-28a(+)-LL-37 (with His-tag) is transformed to Escherichia coli BL21(DE3). The E. coli strain is cultured in LB medium containing 10 μg/mL kanamycin.
2) When the optical density of the cultured bacteria reached approximately 0.6, IPTG was added to the final concentration 50mM. And the bacteria were induced at 18℃ overnight. The harvested bacteria are resuspended with a binding buffer (Sangon Biotech, Shanghai, China), and then the bacteria are lysed by ultrasonication. Purification is performed following the instructions of Ni-NTA SefinoseTM Resin (Sangon Biotech, Shanghai, China).
3) Salmonella Typhimurium SL1344 is cultured in LB medium at 37℃ overnight. Resuspend the cultured S. Typhimurium SL1344 by PBS. All activity tests are implemented in PBS resuspended S. Typhimurium SL1344. Culture the PBS resuspended S. Typhimurium SL1344 to OD600 = 0.35.
4) In the 96-well plates, 20 μl LL-37 solution is added to the final concentration of 8 μg/mL with 180 μl bacteria suspension in each well. Together with 20 μl Elution buffer (Sangon Biotech, Shanghai, China) added with 180 μl bacteria suspension serve as control groups. Each with three biological repeats.
5) Use the Microplate Reader to measure OD600 of the 96-well-plate for 2.5 hours.
6) All experiments above are carried out in 3 biological replications.
Results and analysis
Compared with the control groups, LL-37 do have antibacterial effect with the OD600 has an obvious lower increase. We successfully characterized LL-37 and demonstrated its ability to kill S. Typhimurium SL1344. The results of our experiment are shown in the figure below (Figure 1).

Improvement by 2022 Shanghai_city
Our composite part BBa_K4248005 was improved based on the existing part BBa_K1162006. There is a mini-review of the development of LL-37 protein-related parts. In 2009, group iGEM09_Slovenia firstly designed a basic part BBa_K245114, LL37. In 2012, group iGEM12_Trieste designed the part LL37 (BBa_K875009) using Assembly Standard #10 for both the prefix and suffix and optimizing the coding sequence for E.coli. In 2013, group iGEM13_Utah_State further improved the part LL37 (BBa_K1162006) using Assembly Standard #23 and by removing the stop codon at the end of the coding region. In 2019, group Jilin_China 2019 characterized this part to kill C. Albicans. In 2021, group HZAU-China 2021 characterized this part to kill Salmonella Typhimurium SL1344. Today, our team further improved the LL-37 by adding fusing a new peptide Sparamosin26-54 with a Linker between the two APMs. Furthermore, the codon has been optimized for E. coli to develop our new composite part BBa_K4248005.
In order to prove the function of our new fusion part LL37-Linker- Sparamosin26-54 , we expressed and purified the Fusion AMP, LL-37 AMP, and Sparamosin26-54 AMP, then detected the inhibition in mixed bacteria acquired from Haichang Ocean Park. As the result shown above, our Fusion AMP has achieved nearly twice bacteriostatic ability as efficiency as that of LL-37 peptide. Besides, our project aimed to clean fish tanks. And Fusion AMP could well inhibit mixed bacterial growth in fish tanks, which is in accord with our initial expectation.
LL-37-Linker-Sparamosin26-54
Contribution
We are the Pepsick iGEM team dedicated to developing antimicrobial peptide products for cleaning fish tanks. In order to carry forward the spirit of iGEM, and inherit and spread the value of iGEM, we specially searched the iGEM Biological Parts library for related antimicrobial peptides and picked BBa_K1162002, Spheniscin-2 antimicrobial peptide from the king penguin (Aptenodytes patagonicus). This is a biological part submitted by iGEM13_Utah_State in 2013, with only DNA sequence information and simple text description information. Our team carried out a comprehensive characterization of this part in the laboratory, adding data from antibacterial testing to dedicate its function and properties in inhibiting bacterial growth. This information can be a good reference for future iGEM teams working on antimicrobial peptides.
What's more, we have added a new fusion antimicrobial peptide, which is formed by fusing LL-37 and Sparamosin26-54 through a protein linker, which can effectively combine the antibacterial functions of LL-37 and Sparamosin26-54, not only inhibiting bacterial growth but also inhibiting fungal growth, that is an original and new form of fusion antimicrobial peptides, which also provides more ideas for future iGEM teams to optimize antimicrobial peptide products.
Engineering Success
Construction of expression plasmids
We design the plasmid: a fusion AMP that connects LL-37 and Sparamosin26-54 through a linker, which was also inserted into the pET-28a(+) vector for protein expression, as shown in Figure 1.
To build the plasmid, we let the synthetic company synthesize the DNA fragment of Fusion and integrate it into the pET-28a(+) vector. The returned sequencing comparison results showed that there were no mutations in the ORF region (Figure 2.). Thus, the expression plasmids were successfully constructed. And the last step was extracting the recombinant plasmids from E.coli DH5α and transferring them into E.coli BL21(DE3) competent, so that can be used to express AMP proteins.
Protein expression and purification
In order to obtain the AMPs’ proteins, we expanded the culture in the LB medium and added IPTG to induce protein expression when the OD600 reached 0.4. After overnight induction and culture, we collected the cells and ultrasonic fragmentation of cells to release the intracellular proteins. Next, we used nickel column purification to purify the antibacterial peptide protein we wanted. The concentration of protein was measured as: 0.431mg/mL Fusion. At this point, we got the AMPs’ protein solutions we wanted.
Functional test
To confirm the function of the AMPs to inhibit bacterial growth, we used the real mixed bacteria acquired from Haichang Ocean Park as bacteria, and antibiotics as a positive control for bacteriostatic test experiments.To better show the relationship between the concentration of antimicrobial peptides and the inhibition of bacterial growth, we added 100 μL of mixed bacteria and 100 μL of different concentrations of the AMPs to each tube and repeated them three times for each concentration to form the average data graph with error bars.
As we have seen in Figure 4, the Fusion peptide’s performance (Figure 4C) at each concentration was almost always the best of the three. Furthermore, Fusion widens the gap when the concentration reaches 15 μm. The Fusion peptide did nearly twice as efficient as the rest of the two peptides (Figure 4A, 4B) in the concentration of 15μm, 20μm, and 25μm, proving the success of using the Fusion peptide to enhance our performance of the AMPs.
BNDS-China 2023
Our team intends to utilize LL-37 as a part of our sterilization system targeting S.aureus.
Characterization of sterilization effect against S.aureus
Cell spreading under different concentrations of LL-37
To visualize the bactericidal ability of LL-37, four groups of S. aureus are spread on four chromogenic agars. Dissolved LL-37(1mg/ml) was diluted into different concentrations (1.0mg/ml, 0.5mg/ml) and added into two different chromogenic agars, 200μl for each. 200μl water and 200μl Glacial Acetic Acid were added to the other 2 agars to act as negative and positive control groups.
Figure 1. Overnight S.aureus chromogenic plate of S.aureus treated with different dissolved LL-37 concentrations
A)Glacial Acetic Acid
B)Water
C)0.5mg/ml LL-37
D)1.0mg/ml LL-37
After overnight incubation under 37 °C, the colonies’ density indicates that the higher the concentration of LL-37 is, the more limited S. aureus spread in the agar, which gives us an overview of bactericidal activity of LL-37.
Examining sterilization effect of LL-37 using spectrophotometer(OD 600)
To gain a detailed view of the sterilization effect of LL-37 against S.aureus. at different concentrations, S. aureus is shaken overnight and tested for the original OD 600 value. After the OD 600 value of cultures is close to 1.0, they are separated into 4 different sterilized tubes, 5 ml each. Then 100μl of Glacial Acetic Acid and dissolved LL-37(1mg/ml) is added to 4 tubes respectively, achieving different final concentration (0.1mg/ml, 0.05mg/ml, 0.01mg/ml). Then OD 600 values over time of all 4 tubes are recorded after 0.5min, 5min, 10min, 20min, 30min being treated, each group needs to be repeated three times.
Figure 2. OD600 of S. aureus under different concentrations of LL-37 with respect to time error bars are too small to be visible)
The data clearly indicates the sterilization ability of LL-37. As the time increases, there is an obvious decrease in OD 600 values of S. aureus. At the beginning of the experiment, OD 600 values for all groups is close to 1.0, while the number drops to 0.8 for most groups and even 0.3 for 0.1mg/ml group after 30 minutes. The effect of ll-37 at concentration of 0.01mg/mL is not obvious since its concentration is too low for LL-37 to work normally.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| n/a | LL-37 antimicrobial peptide from Humans (Homo sapiens) |