Part:BBa_K3264013
2Rep-sfGFP
This part is improved from <a href="https://parts.igem.org/Part:BBa_I746916">BBa_I746916</a> We here use sfGFP combined with the an extensive repetitive region-Rep. 2Rep-sfGFP is the recombinant chromoprotein that can mix with the NT-2Rep-CT spidroin in a certain ratio to be artificially spun into the continuous and high function spider silk with green fluorescence property. This part is in the part collection where we provide improved recombinant chromoproteins and spidroins based on the functions of different regions of naturally occurring spidroin. By carrying out different mixing of the proteins of different combinations, we can produce artificially spun silk of different colors and properties.
The part collection includes: Parts that are different kinds of spidroins for silk spining. BBa_K3264000 BBa_K3264005 BBa_K3264007 BBa_K3264009 BBa_K3264018. BBa_K3264019. Parts that for combining with the spidroins to form color silks BBa_K3264012. BBa_K3264013. BBa_K3264014. BBa_K3264015. BBa_K3264016. BBa_K3264017. Part for adding more extensive repetitive region to the spidroin protein BBa_K3264011.
Our part collection can help and inspire future teams to further design recombinant spider silks of more variety and give silks diversified color and property to fulfill the collection.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 292
Usage and Biology
2Rep-sfGFP is the recombinant chromoprotein that can mix with the NT-2Rep-CT spidroin in a certain ratio to be artificially spun into the continuous and high function spider silk with green fluorescence property. This year, we synthesized the part NT-2Rep-CT, to for artificial silk spinning, is the common architecture in two main types of spider silk proteins: major ambulate spidorins(MaSps) and minor ampullate spidorins (MiSps): A non-repetitive N-terminal domain (NT), as well as the C-terminal domain (CT). In between them is an extensive repetitive region (Rep). We believe this part can realize the approach of producing recombinant spidroins (silk proteins) from other chassis and spin them into silk to fulfill the demand of spider silks. However, this part is only responsible for to form continuous silks that are colorless.
Inspired by the functions of 3 main regions in NT-2Rep-CT spidroin and in order to reach a larger range of functions, and apply the artificial spun silk to a wider range of industries such as the clothes industry. This year, we also constructed a part collection of recombinant chromoprotein based on three previous parts: sfGFP (BBa_I746916), eforRed (BBa_K592012), and amilCP(BBa_K592009). Two kinds of improvements are applied to the original chromoprotein: adding a 2Rep region, or adding NT domain, followed by 2Rep region in the front of the nucleotide sequence of chromoprotein.
With the hope that 2Rep can assist the NT-2Rep-CT extensive region’s transition into beta-sheet formation as the 2Rep regions can lie parallel to each other and be formed into beta-sheet structure cohesively. NT’s high solubility in neutral pH and improve it’s blend with NT-2Rep-CT, hence a more continuous and uniformly distributed silk can be artificially spun.
In this part, we added the 2Rep region in front of chromoprotein sfGFP (BBa_xxxxxxx) to form the recombinant chromoprotein.
Reference
[1] Andersson, Marlene, et al. “Biomimetic Spinning of Artificial Spider Silk from a Chimeric Minispidroin.” /Nature Chemical Biology/, vol. 13, no. 3, Sept. 2017, pp. 262–264., doi:10.1038/nchembio.2269.
Characterization
We aimed to use a synthetic biology approach to combine a standardized DNA part 2Rep-sfGFP, and transform constructs to metabolically engineered Escherichia coli (E.coli) for bioproduction.
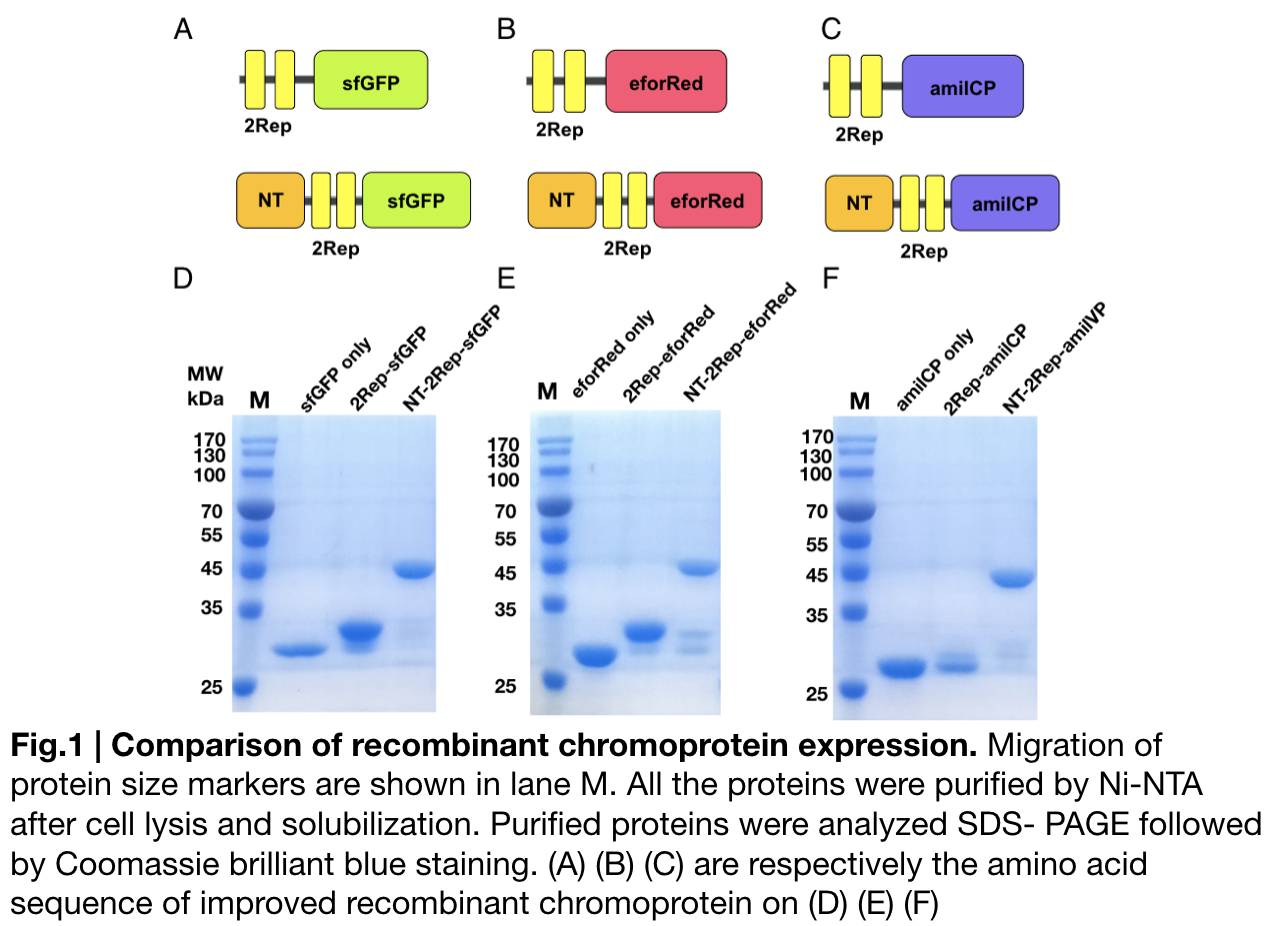
Purification and SDS PAGE
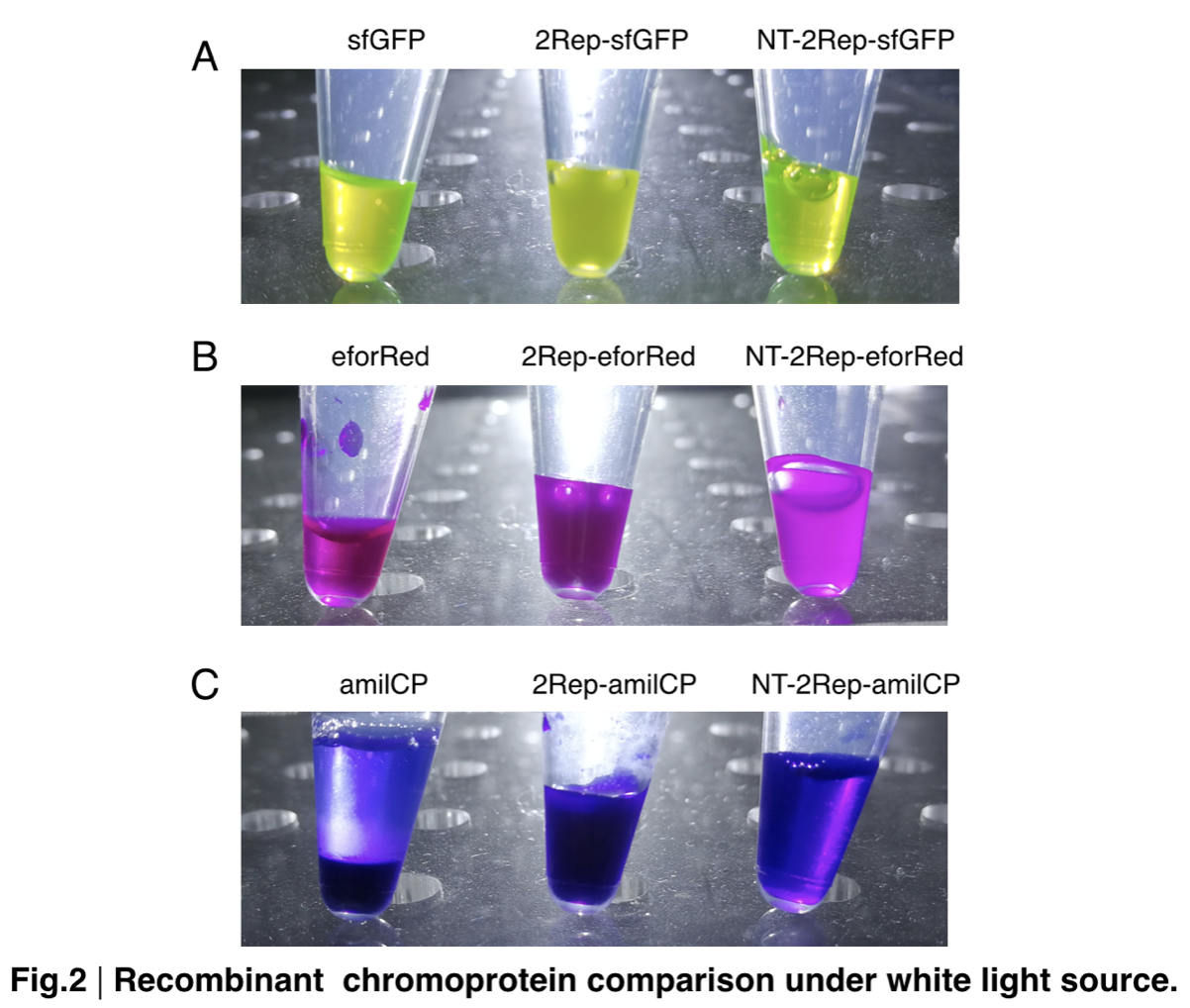
In order to detect whether protein expression was induced by adding isopropylthiogalactoside (final concentration 0.3mM), we used SDS-page(10%) to determine the presence of target protein. Target protein after purification by Ni-NTA method was compared to their original chromoprotein only. Results showing the size chromoprotein only, 2Rep-chromoprotein, and NT-2Rep-chromoprotein are in an increasing trend as regions are added, this result is constant in all three of our chromoprotein. This suggests that our 2Rep protein domain or NT-2Rep region is successfully added on to our pure chromoprotein and well induced. Placing the three kinds of chromoprotein, each of them with two kinds of improvements to compare. All 2Rep-chromoprotein under white light source have the most opacity in their own group of chromoprotein of same color. All NT-2Rep-chromoprotein is translucence. However, whereas sfGFP and eforRed chromoprotein only are relatively transparent to all other in their roll, amilCP chromoprotein only self-assembles and gradually subside. The precipitates of amilCP chromoprotein can be dissolved into the solution again after a small shake but reforms in a short period of time.
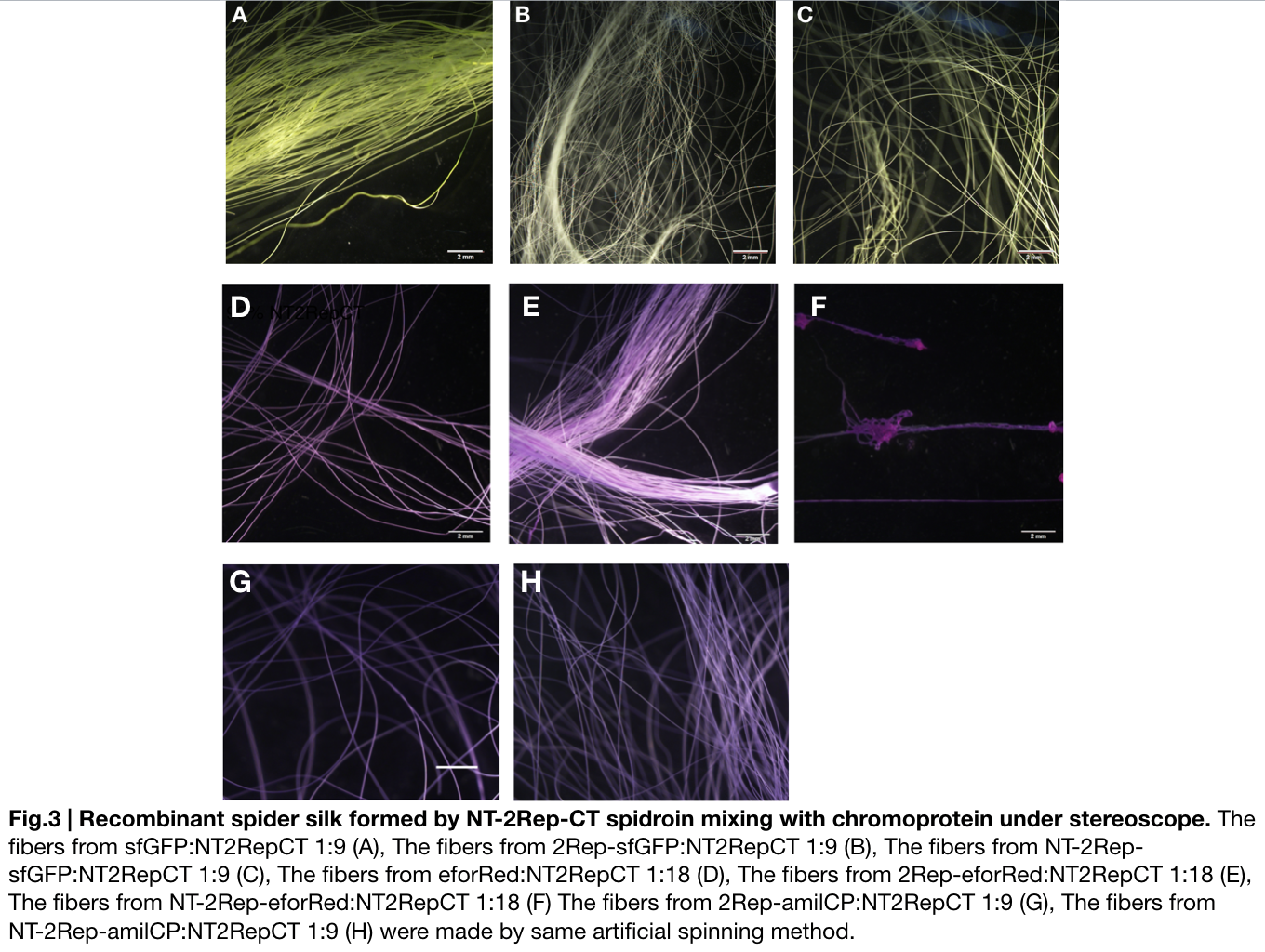
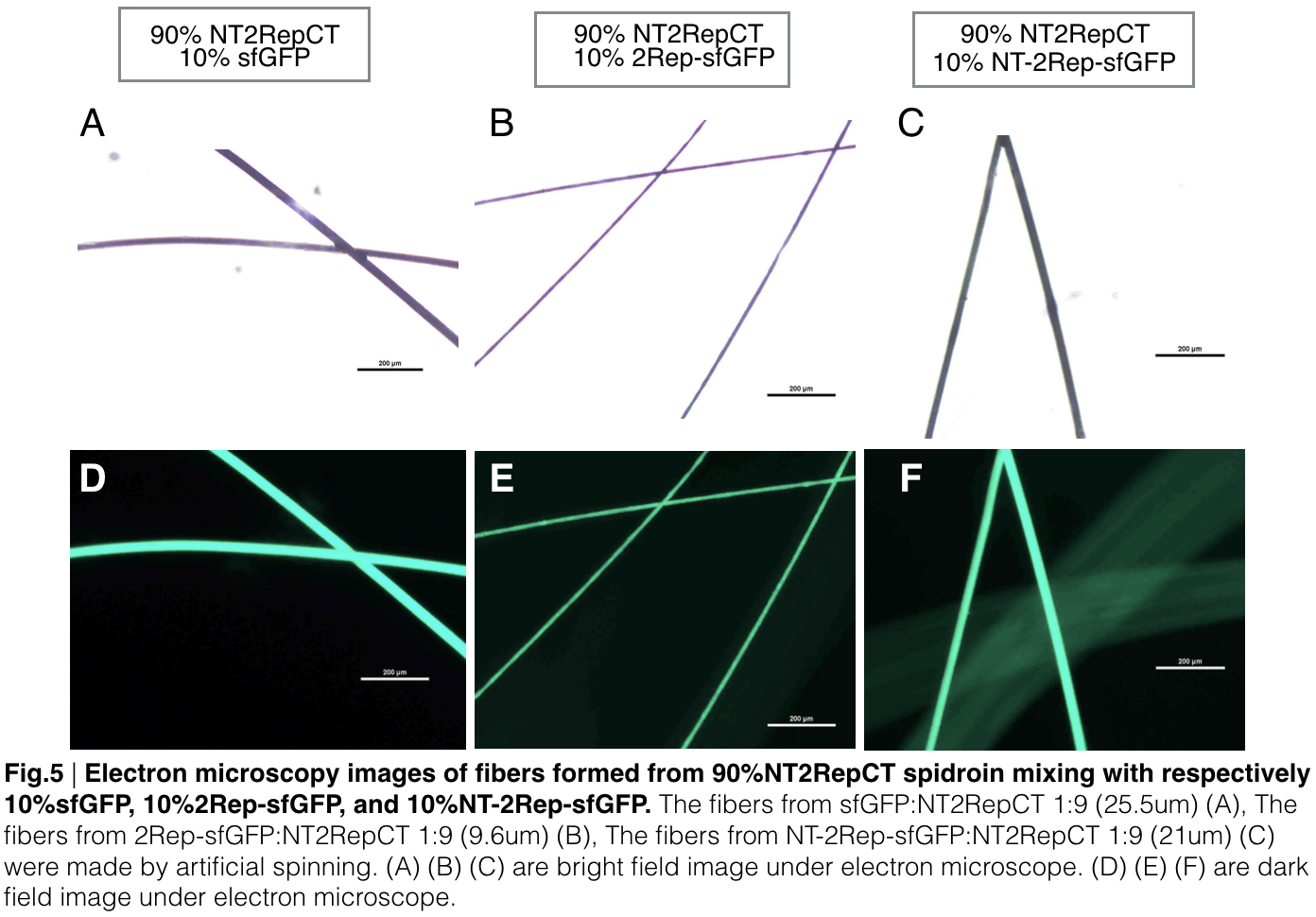
Fiber spinning of NT2RepCT with recombinant chromoprotein
Recombinant chromoprotein 2Rep-chromoprotein concocting with NT-2Rep-CT spidroin in the suitable ratio can form the most continuous and stable silk when spinning into 100%isopropanol comparing with the silk spun by replacing 2Rep-chromoprotein with chromoprotein only or NT-2Rep-chromoprotein. It’s also the most transmittance The stable feature of this particular silk spun supports our assumption to let the 2Rep region bounded to the chromoprotein assist the NT-2Rep-CT extensive region’s transition into beta-sheet formation as the 2Rep regions can lie parallel to each other and be formed into a beta-sheet structure cohesively without disruption to the dimer forming at NT or the amyloid-like fibril forming at CT when carrying out artificial silk spinning at low pH or in 100% isopropanol. When NT-2Rep is added in front of chromoprotein instead of 2Rep, and mixed with NT-2Rep-CT in the same ratio to carry out artificial spinning, the liquidity of the of proportion spinning into 100% isopropanol is higher than the other two, it supports our assumption that NT’s high solubility in neutral pH and improve it’s blend with NT-2Rep-CT, hence a more continuous and uniformly distributed silk can be artificially spun.
Due to the easily self-assemble property of amilCP chromoprotein, we were unable to artificially spin its mix with NT-2Rep-CT spidroin into continuous silk, become the syringe head was stuck immediately after the dose of mixture enters the syringe and amilCP starts to self-assemble and subside to the needle head.
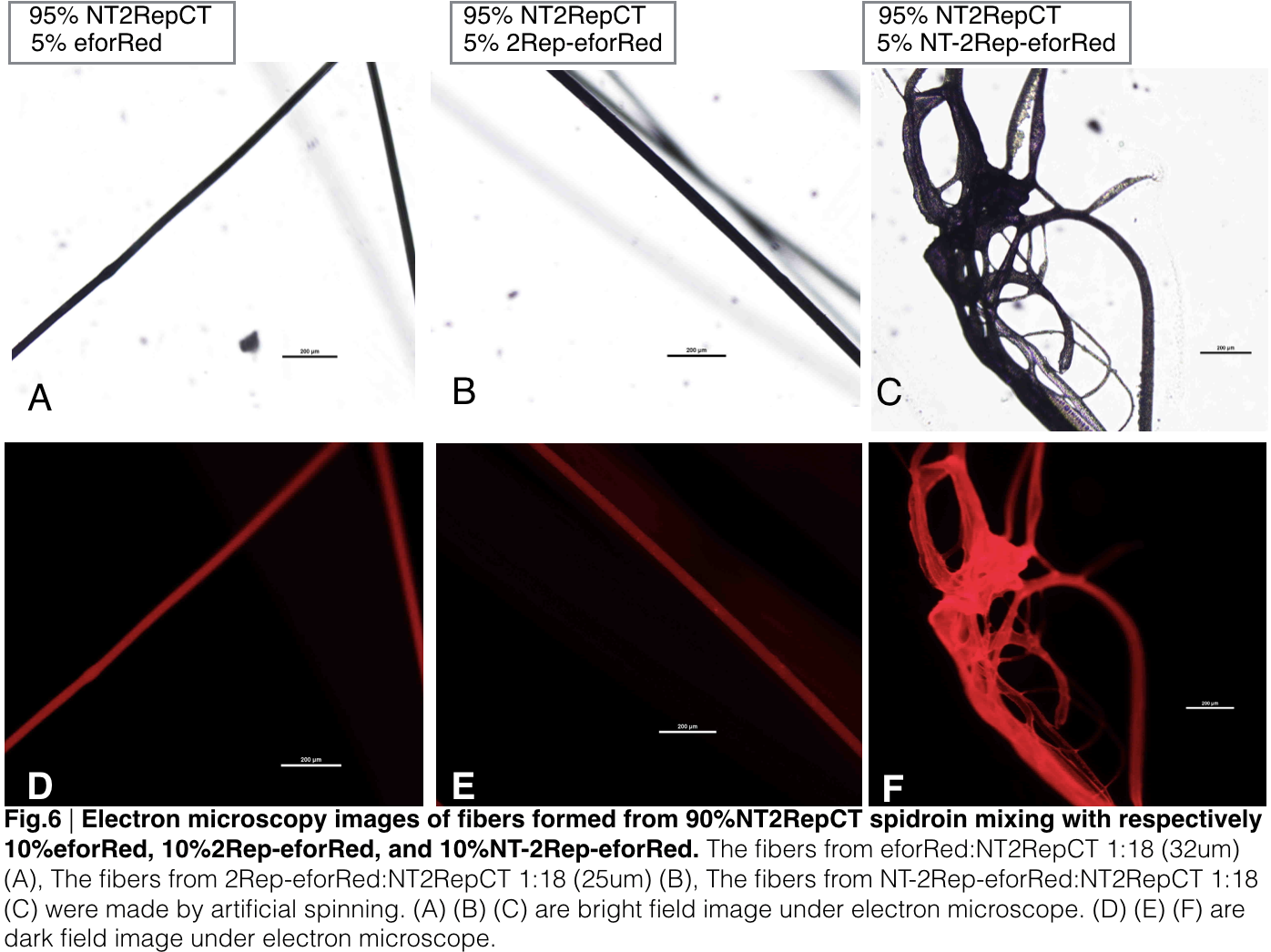
When we were trying to adjust the suitable ratio of eforRed recombinant chromoprotein (eforRed only, 2Rep-eforRed, and NT-2Rep-eforRed), we found all of them were unable to be artificially spin with NT-2Rep-CT spidroin when their concentration in the whole mix is higher than 5%. We successfully spun silk with eforRed only and 2Rep-eforRed that had concentration of 5% in the entire mix, but only discontinuous silks are formed by the mix of NT-2Rep-eforRed of the same concentration.
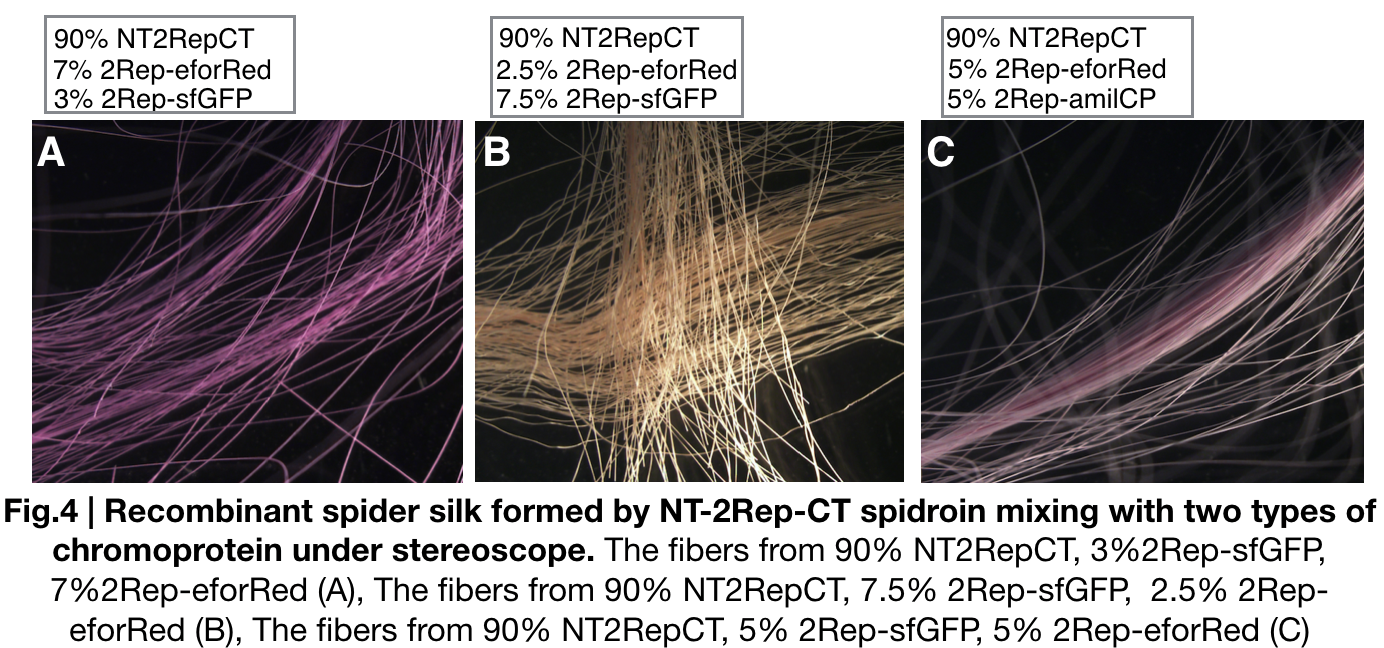
We further investigated the feasibility to mix two kinds of recombinant chromoprotein: 2Rep-chromoprotein, into the NT-2Rep-CT spidroin, taking up in tota 10% of the entire mix, to artificially spin into more colorful and stable silk. The result has shown to us that 2Rep-sfGFP can add brightness feature to the silks. Also, the concentration of 2Rep-eforRed can be raised to 7.5% and still continuously form silks.
Scanning electron microscopy of fibers
Aiming to verify the distribution uniformity of the mixture of recombinant chromoprotein and spidroin after spinning into fiber, we looked at our artificial spun silks under electron microscope. AmilCP is not scanned due to its lack of fluorescence. We randomly selected the cross section in our continuous silk, it has clearly shown that the fiber circularity is qualified as a uniformly distributed silk. For both sfGFP and eforRed, when 2Rep is added to the front of chromoprotein, the silk formed is has the smallest diameter among all the silks in the chromoprotein same group.
| None |