Part:BBa_K3110007
Medium Promoter Strong RBS lldD
This construct has L-Lactate Dehydrogenase (lldD) under the control of a medium promoter and a strong RBS.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1202
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 639
Usage and Biology
lldD cleaves L-lactate to pyruvate. Our intention of using this part is to have control over the detection threshold of L-Lactate by the lldPRD regulatory region. We have also designed other parts which produce various higher and lower concentrations of lldD by varying the strength of the promoter and RBS controlling its production.
Characterization of the Part
Experimental Procedure
- We chose BL21(DE3) as our chassis and transformed both the plasmids in BL21(DE3) separately. Cultured them in M9-Glycerol(with Chloramphenicol of concentration 34 ng/ul) for 14-16 hrs at 37C, 200 r.p.m.(Primary Culture). After that, we put the secondary culture of both at an OD of O.1 in M9-Glycerol medium (With Chloramphenicol concentration- 34 ng/ul) and kept at 37C for 5-6 hours.
- Once the OD of the secondary culture reached 0.4, we transferred the culture to 96 well microtiterplate with appropriate blank (M9 + required lactate concentration) after induction with different concentrations of lactate (0 mM, 0.01 mM, 0.03 mM, 0.06 mM, 0.1 mM, 0.3 mM, 0.6 mM, 1 mM, 3 mM, 6 mM, 10 mM, 13 mM, 16 mM, 20 mM) and kept in plate reader. Our assays were done in technical triplicates and repeated thrice with different biological samples.
- We used the Gen5 software to program the plate reader. The cultures were kept at 37degC with orbital shaking at 282cpm for 27:30 mins. This was followed by reading OD (at 600 nm) and then fluorescence at 480 excitation and 510 emission with 70 gain. This was iterated over cycles set for 8hrs.
- Analysed the data after completion of the program.
Corrected Fluorescence = Fluorescence (Sample) - Fluorescence (Blank) Corrected OD = OD (Sample) - OD (Blank) Normalized Fluorescence = Corrected Fluorescence / Corrected OD Each experimental data set was fitted to a Hill function (using origin).
First we chose only lldRO1-J23117-lldRO2 Strong RBS sfGFP to distinguish between cancerous and non-cancerous micro-environments.

From the above graph it's clear that the construct lldRO1-J23117-lldRO2 Strong RBS sfGFP (Part:BBa_K3110040) gives a sigmoidal pattern for detection of lactate with a threshold of around 0.45 mM. However, this is not an appropriate threshold since the construct might fail to distinguish Cancerous vs Non-cancerous microenvironments, both of which have lactate concentration than 0.45mM. Hence we have intended to use this construct in combination with our library of four gene constructs: lldD, lldP+lldD, lldR+lldD and lldP+lldR+lldD under promoter and RBS of different strengths.
We hypothesized that, if we can increase the amount of Lactate Dehydrogenase inside the cell, it will cleave some amount of Lactate. So, the surge in sfGFP expression should be seen in a higher concentration of lactate induction.
Genetic Design for the Characterization
So, we designed the following gene construct to produce more LldD (L-Lactate dehydrogenase) in a different plasmid (pSB3K3): <bold>Medium Promoter-Strong RBS-lldD-Terminator</bold> We used medium promoter and cloned into a low copy number plasmid so that the amount of lldD inside the cell shouldn’t be too high to cleave all the lactate inside the cell and there is no lactate left inside the cell to induce O1PO2. Also, pSB3K3 and pSB1C3 are compatible plasmids and hence can be co-transformed. Then we co-transformed the 3 constructs in BL21(DE3) in 2 different combinations:
- Medium P-Strong RBS-lldD (in pSB3K3) + O1PO2-Strong RBS-sfGFP (in pSB1C3)
- Medium P-Strong RBS-lldD (in pSB3K3) + P-Strong RBS-sfGFP (in pSB1C3)
Results
We chose 480 min for further analysis.
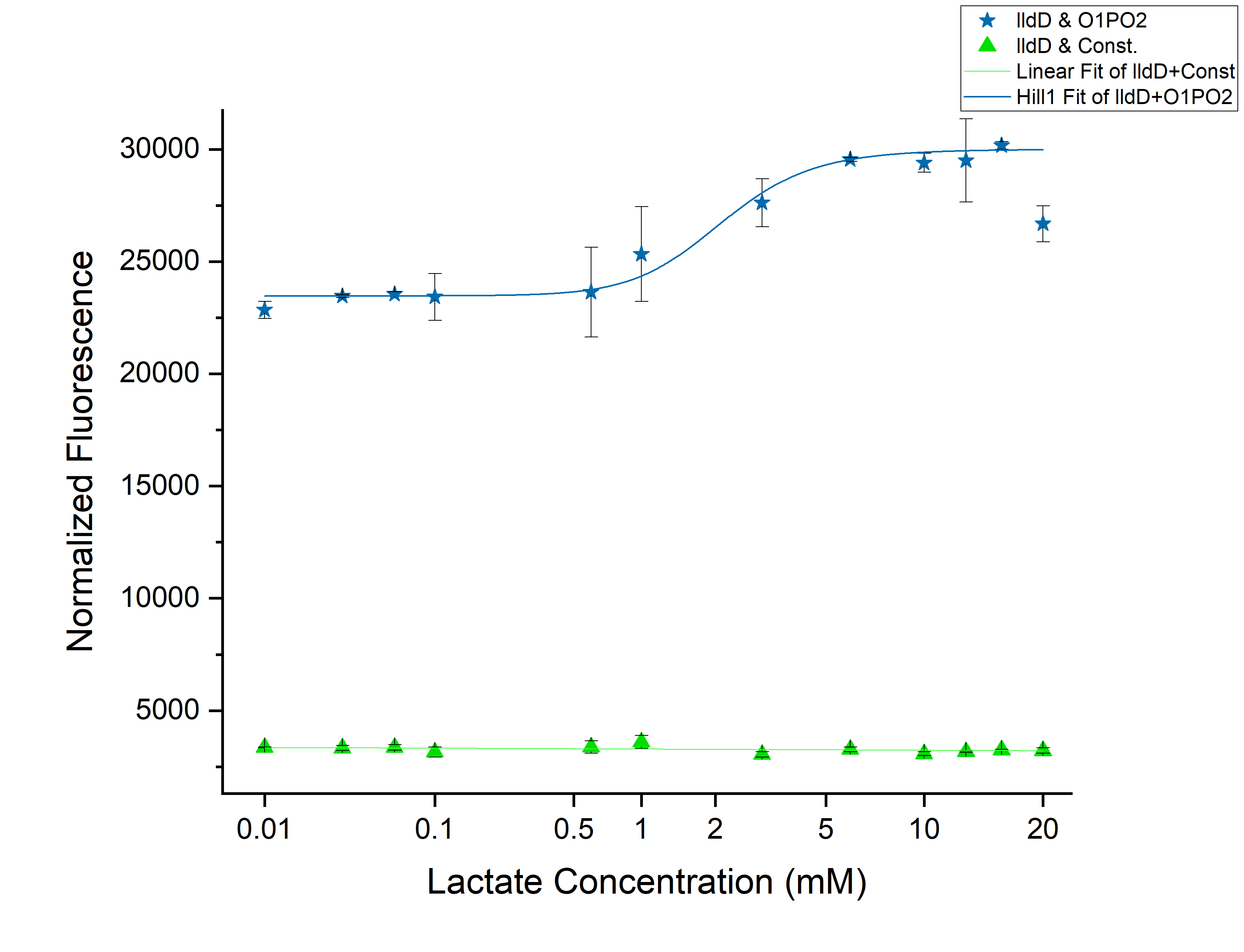
From the above graph, we observed an increase in threshold to 2 mM as we expected. Although this might not be able to differentiate cancer from non-cancerous cells. However, an increase of threshold suggests that our idea of using lldD as the modulator will work. To clearly understand the effect of ectopic lldD on O1PO2, we plotted the graphs for only O1PO2-Strong-RBS-sfGFP and O1PO2-Strong-RBS-sfGFP along with lldD together.
Comparison between only O1PO2 and O1PO2 along with lldD
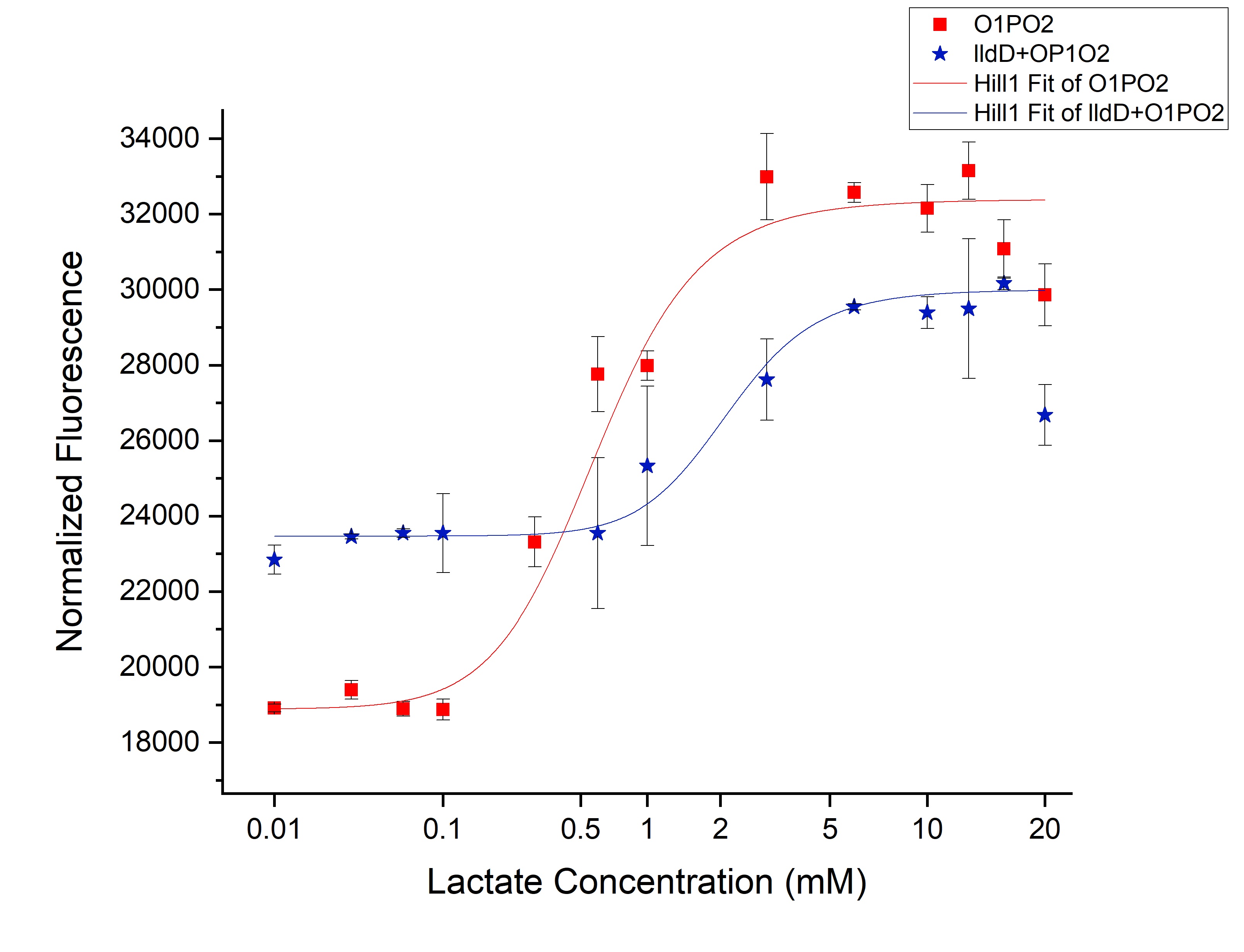
From this picture, we can clearly see an increase in threshold from 0.45 mM (without lldD) to 2 mM (with lldD) as we expected. Another, major drawback that we observed from this was a decrease in sensitivity i.e. even at sub-threshold lactate levels, the fluorescence observed in the presence of lldD was high. Not only this, at the aforementioned threshold levels, the intensity was lower than in the case without lldD. This could be rectified by using lldP and lldR along with lldD.</p>
| None |

