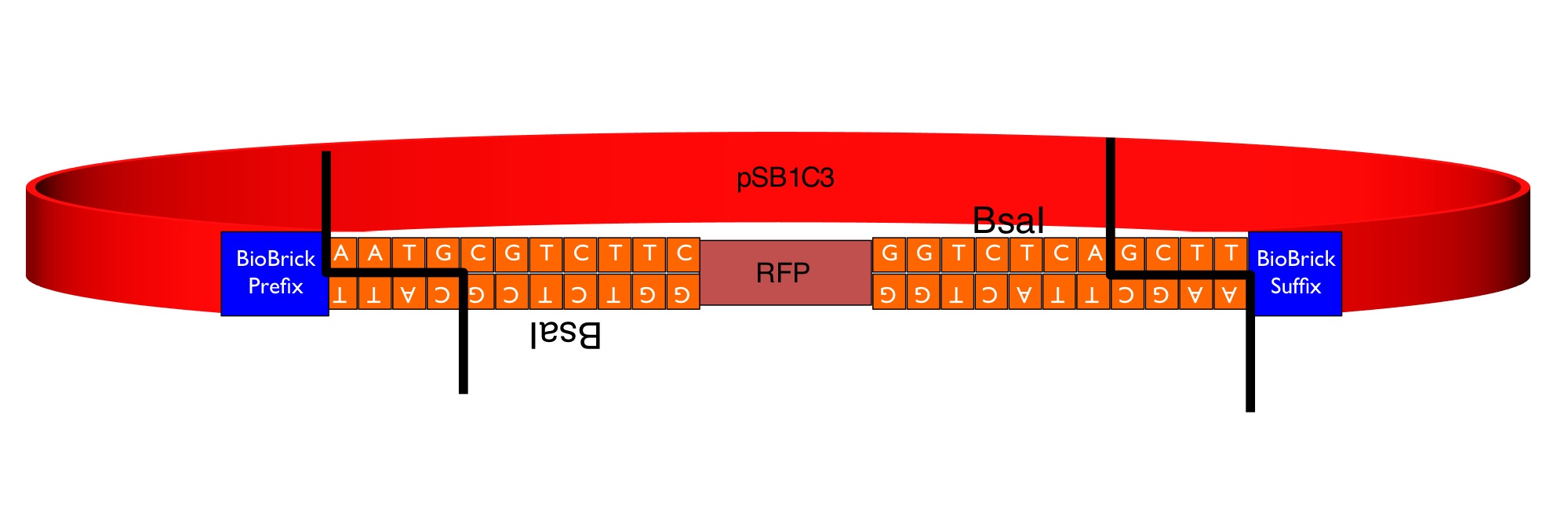
Part:BBa_K1467200
RFP coding device - Golden Gate Module Flipper
This part is a "GoldenGate Flipper" that can be used to convert CDS1 (coding sequence) modules from the GoldenGate MoClo Assembly Standard into standard BioBricks. It consists of the RFP reporter (from BBa_J04450) flanked by an inverted pair of BsaI recognition sequences. GoldenGate MoClo parts can be flipped into BrioBrick parts in a one-pot, one-step, digestion-ligation GoldenGate cloning reaction. The image below shows the sequences inserted (in orange) between the RFP and the BioBrick prefix and suffix that enable the flipper to accept MoClo CDS1 parts, which begin AATG and end GCTT.
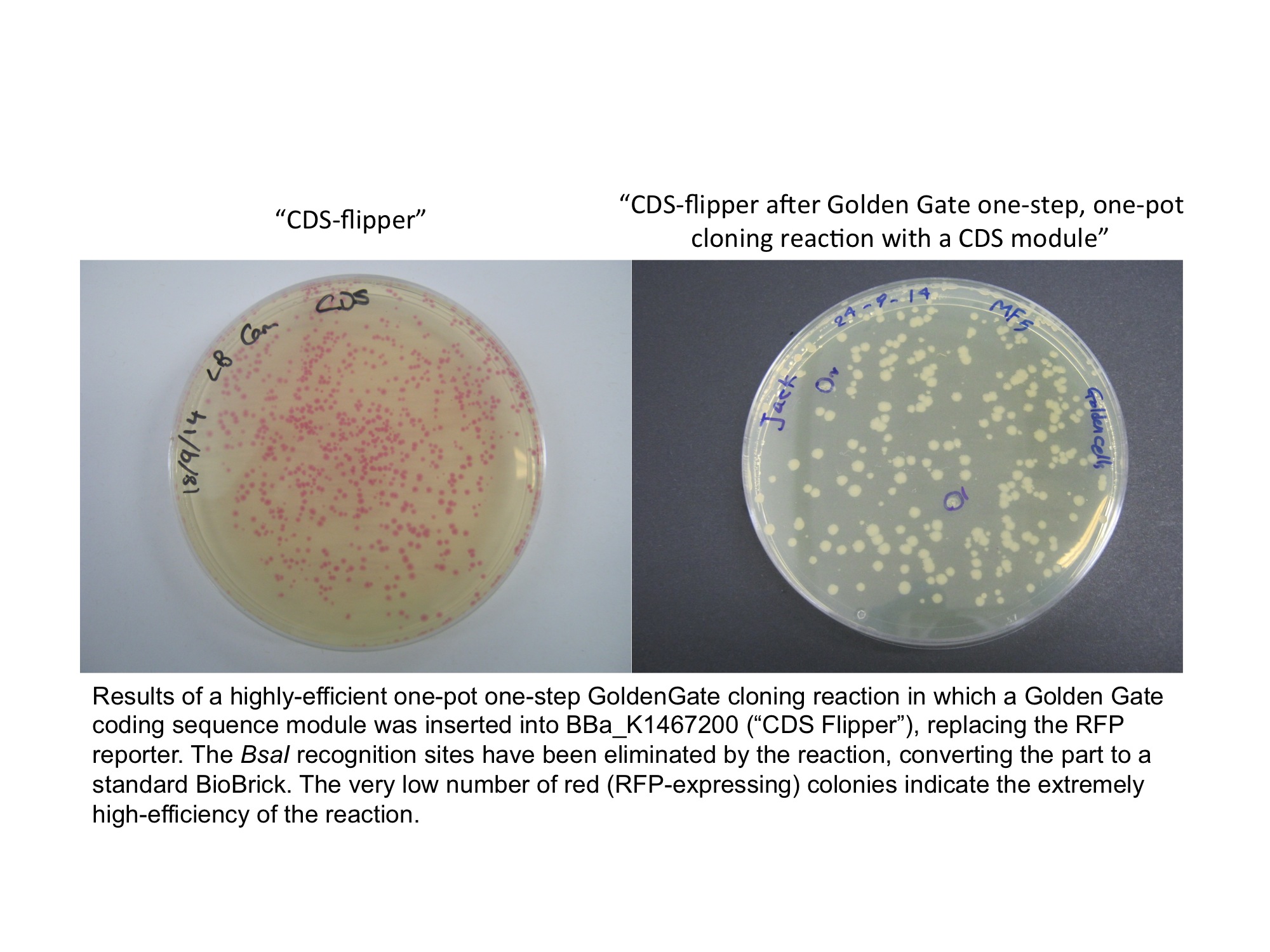
The colonies containing the RFP part are clearly red in color under natural light after about 18 hours. Smaller colonies are visibly red under UV. The RFP part does not contain a degradation tag and the RBS is strong. When the flipper reaction is successful the colonies become white as the RFP sequence is replaced by the new part. No part of the BsaI recognition sequence will remain between the BioBrick suffix and the new part.
iGEM14_NRP-UEA-Norwich have used this part to convert GoldenGate "CDS" (coding sequence) modules from the GoldenGate MoClo Assembly Standard into standard BioBricks. The cloning reaction was a Golden Gate one-step, one-pot reaction where the restriction enzyme, ligase and (intact, uncut) donating and recipient plasmids were all incubated together in a single reaction before transformation into competent E.coli cells. Cloning was very successful as the vast majority of the colonies are white as the RFP sequence was replaced by the new part.
Contribution - Calgary 2019
Group: [http://2019.igem.org/Team:Calgary iGEM Calgary 2019]
Author: Sravya Kakumanu, Sara Far, Nimaya De Silva, Cassandra Sillner
Summary: We improved this part by substituting a strong constitutive promoter (BBa_J23100) for the LacI-regulated promoter. We also exchanged the RBS for a stronger one, and codon optimized the part for E. coli. These changes were aimed to reduce the incubation time required for red colonies to become visible under normal light.
Documentation: iGEM Calgary used the improved version of this part to convert pSB1A3 to our Golden Gate destination vector.
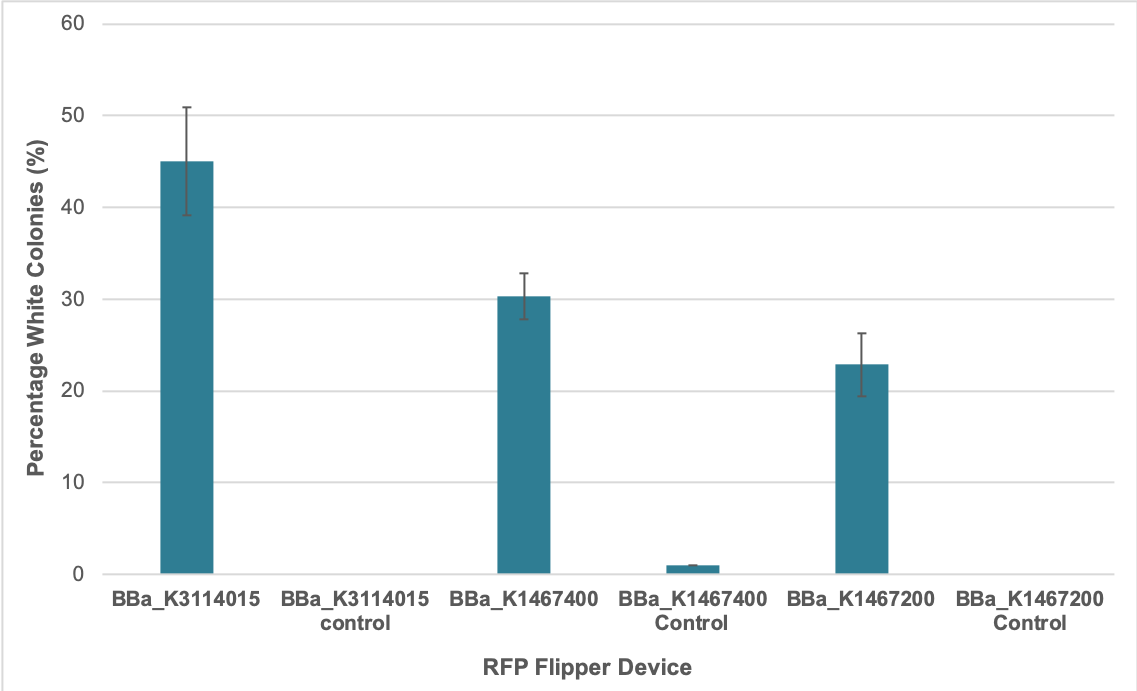
We determined the average percent successful assembly for our improved part BBa_K3114015 as well as the original BBa_K1467400 based on the proportion of white colonies following transformation of Golden Gate assembly samples. Golden Gate assembly reactions were set up to each contain the same concentration of destination vector and the required DNA inserts in the form of PCR products. A 1:1 insert:destination vector ratio was used in each reaction. They were conducted as per our Golden Gate assembly protocol. BBa_K3114015 and BBa_K1467400 were used in four-insert assembly reactions using parts:
The other RFP Golden Gate flipper (BBa_K1467200) that we characterized was used in two-insert assembly reactions using parts:
The Golden Gate assembly products were transformed into E. coli DH5-alpha. Some colonies became visibly red under normal light after 12 hours for the improved part BBa_K3114015 and 18 hours for BBa_K1467400 and the other RFP device Golden Gate flipper (BBa_K1467200) that we characterized. 24 hours after transformation, we counted the number of red versus white colonies for each of the replicates. The results suggest that using the improved RFP flipper BBa_K3114015 resulted in a higher percentage of successful assembly reactions compared to the original RFP flipper BBa_K1467400. However, the mechanism behind this is unclear.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 793
Illegal AgeI site found at 905 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1082
Illegal BsaI.rc site found at 6
| None |